Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases - PubMed (original) (raw)
Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases
Adrian A Cuenca et al. Development. 2003 Apr.
Abstract
Polarization of the C. elegans zygote along the anterior-posterior axis depends on cortically enriched (PAR) and cytoplasmic (MEX-5/6) proteins, which function together to localize determinants (e.g. PIE-1) in response to a polarizing cue associated with the sperm asters. Using time-lapse microscopy and GFP fusions, we have analyzed the localization dynamics of PAR-2, PAR-6, MEX-5, MEX-6 and PIE-1 in wild-type and mutant embryos. These studies reveal that polarization involves two genetically and temporally distinct phases. During the first phase (establishment), the sperm asters at one end of the embryo exclude the PAR-3/PAR-6/PKC3 complex from the nearby cortex, allowing the ring finger protein PAR-2 to accumulate in an expanding 'posterior' domain. Onset of the establishment phase involves the non-muscle myosin NMY-2 and the 14-3-3 protein PAR-5. The kinase PAR-1 and the CCCH finger proteins MEX-5 and MEX-6 also function during the establishment phase in a feedback loop to regulate growth of the posterior domain. The second phase begins after pronuclear meeting, when the sperm asters begin to invade the anterior. During this phase (maintenance), PAR-2 maintains anterior-posterior polarity by excluding the PAR-3/PAR-6/PKC3 complex from the posterior. These findings provide a model for how PAR and MEX proteins convert a transient asymmetry into a stably polarized axis.
Figures
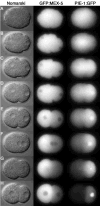
Fig. 1
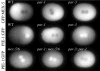
GFP:PAR-2 and GFP:PAR-6 dynamics in wild-type and mutant embryos (Movies 1–2, 10–11, 20–21). (A) Pronuclear formation. (B) Cessation of ruffling in posterior. (C) Pseudocleavage. (D) Pronuclear meeting. (E) Spindle rotation. (F) Mitosis. (G) two-cell stage. Quicktime movies of this and subsequent figures are accessible at ftp://
www.wormbase.org/pub/wormbase/datasets/seydoux\_2003
. C. elegans zygotes are approximately 50 μm in length.
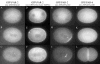
Fig. 2
GFP:PAR-2 and GFP:tubulin dynamics in wild-type embryos. (A–D) An embryo expressing both GFP:PAR-2 and GFP:tubulin (Movie 3). (E–H) An embryo expressing GFP:PAR-2 (Movie 4).
Fig. 3
GFP:MEX-5 and PIE-1:GFP dynamics in wild-type embryos (Movies 5 and 6). (A) Pronuclear formation. (B–D) Pronuclear migration. (D) Pseudocleavage. (F) Pronuclear meeting. (G) Mitosis. (H) Two-cell stage. Note that PIE-1:GFP accumulates in the male pronucleus during pronuclear migration, on the posterior centrosome during mitosis, and in the P1 nucleus in the two-cell stage. PIE-1:GFP and GFP:MEX-5 also accumulate on the cytoplasmic P granules (best seen in the movies).
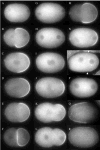
Fig. 4
GFP:PAR-2 and GFP:PAR-6 dynamics in nmy-2(RNAi) embryos. A,D,G,J are embryos during pronuclear migration; B,E,H,K are at pronuclear meeting; C,F,I are in late mitosis; L is a 2-cell embryo. GFP:PAR-2 remains off the cortex in the most severely affected embryos (AC; Movie 15), unless par-6 is also removed (D-F, Movie 16). In more weakly affected embryos [_nmy-2(RNAiClassII)_], GFP:PAR-2 appears on the cortex around pronuclear meeting (H, Movie 18). Note PAR-2 accumulation in foci around pronuclei and/or spindles (C,H,I). GFP:PAR-6 remains uniformly distributed throughout the cortex even in the most weakly affected embryos (J–L, Movie 17).
Fig. 5
PAR-1 and MEX-5/6 function in a feedback loop to regulate expansion of the posterior domain. (A–F) par-1(RNAi) zygote expressing GFP:PAR-2 (Movie 22). (A) Pronuclear formation. (B) Pronuclear migration. (C) Pronuclear meeting. (D–E) Mitosis. (F) Two-cell stage. The GFP:PAR-2 domain is expanded at pseudocleavage compared to wild type (see Fig. 1). (G–L) mex-5(RNAi)mex-6(RNAi) zygote expressing GFP:PAR-2 (Movie 28). Stages same as in A–F. GFP:PAR-2 domain expansion is slowed down compared to wild type (see Fig. 1). (M) GFP:PAR-2 in par-1(RNAi) pie-1(RNAi) (Movie 27). Expansion of the PAR-2 domain is not suppressed (compare with B). (N) GFP:PAR-2 in par-1(RNAi) mex-5(RNAi)mex-6(RNAi) (Movie 26). Expansion of the PAR-2 domain is suppressed. Identical results were obtained in par-1(it51) mex-5(RNAi)mex-6(RNAi) embryos stained for PAR-2 (not shown). (O,P) Wild-type (O) and mex-5(zu199)mex-6(pk440) (P) zygotes stained with PAR-1 antibody. Arrows point to the ends of the PAR-1 domain in the wild-type embryo. PAR-1 is not visible on the cortex of this mex-5(zu199)mex-6(pk440) zygote. (Q,R) Wild-type (Q) and mex-5(zu199)mex-6(pk440) (R) zygotes stained with PAR-3 antibody. PAR-3 extends further posterior.
Fig. 6
Regulation of PIE-1’s nuclear:cytoplasmic ratio by MEX-5/6 is under PAR-1 control. GFP:MEX-5 is primarily cytoplasmic in all genotypes. In contrast, the nuclear:cytoplasmic ratio of PIE-1:GFP varies depending on the genotype as described in text.
Fig. 7
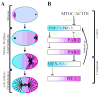
Model for polarization of the C. elegans zygote. (A) Graphic depiction of the establishment and maintenance phases during polarization of the zygote. Proteins that localize to the anterior are in blue (PAR-3, PAR-6, PKC-3 on the cortex, MEX-5 and MEX-6 in the cytoplasm). Proteins that localize to the posterior are in pink (PAR-2 and PAR-1 on the cortex, PIE-1 in the cytoplasm). During meiosis, all are uniformly distributed throughout the zygote (purple color). Circles: pronuclei, Black lines: microtubules. (B) Sequential repression model [modified from Kemphues (Kemphues, 2000)]. Lines with bars depict antagonistic interactions, whereas lines with arrows depict positive interactions. In this model, we show that PAR-1 is restricted to the posterior by the anterior PARs. This hypothesis is supported by the observation that, in late one-cell embryos, PAR-1 is present throughout the cortex in par-3 mutants and par-3;par-2 double mutants (Etemad-Moghadam et al., 1995; Boyd et al., 1996). Further support for this model awaits analysis of PAR-1 dynamics in live embryos.
Similar articles
- The Caenorhabditis elegans par-5 gene encodes a 14-3-3 protein required for cellular asymmetry in the early embryo.
Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ. Morton DG, et al. Dev Biol. 2002 Jan 1;241(1):47-58. doi: 10.1006/dbio.2001.0489. Dev Biol. 2002. PMID: 11784094 - Coupling between cytoplasmic concentration gradients through local control of protein mobility in the Caenorhabditis elegans zygote.
Wu Y, Zhang H, Griffin EE. Wu Y, et al. Mol Biol Cell. 2015 Sep 1;26(17):2963-70. doi: 10.1091/mbc.E15-05-0302. Epub 2015 Jul 8. Mol Biol Cell. 2015. PMID: 26157168 Free PMC article. - Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2.
Zonies S, Motegi F, Hao Y, Seydoux G. Zonies S, et al. Development. 2010 May;137(10):1669-77. doi: 10.1242/dev.045823. Epub 2010 Apr 14. Development. 2010. PMID: 20392744 Free PMC article. - Cell polarity and the cytoskeleton in the Caenorhabditis elegans zygote.
Schneider SQ, Bowerman B. Schneider SQ, et al. Annu Rev Genet. 2003;37:221-49. doi: 10.1146/annurev.genet.37.110801.142443. Annu Rev Genet. 2003. PMID: 14616061 Review. - The PAR network: redundancy and robustness in a symmetry-breaking system.
Motegi F, Seydoux G. Motegi F, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Sep 23;368(1629):20130010. doi: 10.1098/rstb.2013.0010. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24062581 Free PMC article. Review.
Cited by
- Regulation of Microtubule Dynamics in Axon Regeneration: Insights from C. elegans.
Tang NH, Chisholm AD. Tang NH, et al. F1000Res. 2016 Apr 27;5:F1000 Faculty Rev-764. doi: 10.12688/f1000research.8197.1. eCollection 2016. F1000Res. 2016. PMID: 27350865 Free PMC article. Review. - Oligomerization and positive feedback on membrane recruitment encode dynamically stable PAR-3 asymmetries in the C. elegans zygote.
Lang C, Maxian O, Anneken A, Munro E. Lang C, et al. bioRxiv [Preprint]. 2024 Aug 28:2023.08.04.552031. doi: 10.1101/2023.08.04.552031. bioRxiv. 2024. PMID: 39253498 Free PMC article. Preprint. - A genome-wide RNAi screen for enhancers of par mutants reveals new contributors to early embryonic polarity in Caenorhabditis elegans.
Morton DG, Hoose WA, Kemphues KJ. Morton DG, et al. Genetics. 2012 Nov;192(3):929-42. doi: 10.1534/genetics.112.143727. Epub 2012 Aug 10. Genetics. 2012. PMID: 22887819 Free PMC article. - RAB-5 controls the cortical organization and dynamics of PAR proteins to maintain C. elegans early embryonic polarity.
Hyenne V, Tremblay-Boudreault T, Velmurugan R, Grant BD, Loerke D, Labbé JC. Hyenne V, et al. PLoS One. 2012;7(4):e35286. doi: 10.1371/journal.pone.0035286. Epub 2012 Apr 24. PLoS One. 2012. PMID: 22545101 Free PMC article. - Stoichiometric interactions explain spindle dynamics and scaling across 100 million years of nematode evolution.
Farhadifar R, Yu CH, Fabig G, Wu HY, Stein DB, Rockman M, Müller-Reichert T, Shelley MJ, Needleman DJ. Farhadifar R, et al. Elife. 2020 Sep 23;9:e55877. doi: 10.7554/eLife.55877. Elife. 2020. PMID: 32966209 Free PMC article.
References
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. - PubMed
- Doe CQ. Cell polarity: the PARty expands. Nat Cell Biol. 2001;3:E7–E9. - PubMed
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. - PubMed
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases