Collaborative competition mechanism for gene activation in vivo - PubMed (original) (raw)
Collaborative competition mechanism for gene activation in vivo
Joanna A Miller et al. Mol Cell Biol. 2003 Mar.
Abstract
The mechanism by which gene regulatory proteins gain access to their DNA target sites is not known. In vitro, binding is inherently cooperative between arbitrary DNA binding proteins whose target sites are located within the same nucleosome. We refer to such competition-based cooperativity as collaborative competition. Here we show that arbitrarily chosen foreign DNA binding proteins, LexA and Tet repressor, cooperate with an adjacently binding endogenous activator protein, Gcn4, to coactivate expression of chromosomal reporter genes in Saccharomyces cerevisiae. Coactivation requires that the cooperating target sites be within a nucleosome-length distance; it leads to increased occupancy by Gcn4 at its binding site; and it requires both Gcn5 and Swi/Snf which, at an endogenous Gcn4-dependent promoter, act subsequent to Gcn4 binding. These results imply that collaborative competition contributes to gene regulation in vivo. They further imply that, even in the presence of the cell's full wild-type complement of chromatin remodeling factors, competition of regulatory proteins with histone octamer for access to regulatory target sites remains a quantitative determinant of gene expression levels. We speculate that initial target site recognition and binding may occur via spontaneous nucleosomal site exposure, with remodeling factor action required downstream to lock in higher levels of regulatory protein occupancy.
Figures
FIG. 1.
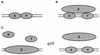
Types of protein-protein cooperativity. (A) Conventional cooperativity. Proteins X and Y touch each other with some favorable free energy. (B) Indirect cooperativity. Proteins X and Y both bind protein Z; thus, the presence of Z confers cooperativity on the binding of X and Y. Examples of such bridging occur for multiple transcription factors in the preinitiation complex (36). Alternatively, a specific Z could bend the DNA between X and Y, enabling conventional cooperative contacts (see text). The HMGI(Y) proteins in the beta interferon enhanceosome illustrate both of these (panels A and B), engaging in direct protein-protein interactions and favorably altering the DNA conformation (25). (C) Competitive cooperativity. Proteins X and Y collaborate in their competition against a common rival, Z. Z may dissociate or change conformation upon binding of X and Y. As an aside, we note that collaborative competition need not be restricted to DNA binding proteins. For example, two proteins could compete with another protein for binding sites on a fourth protein.
FIG. 2.
DNA constructs. Wild type indicates the natural HIS3 upstream region which contains two major transcription start sites (+1 and +13), two TATA boxes (TC and TR), a GRE, and a poly(dA-dT) element. In the LexAop construct, the poly(dA-dT) tract from the HIS3 upstream element has been replaced with the LexA operator (TACTGTATGAGCATACAGTA). The mtGRE construct is identical to LexAop except that the GRE has been mutated (ATCACTCGT; mutations shown in bold). The mtLexAop construct has two point mutations in LexAop (TACCGTATGAGCATACGGTA). In the GFP construct, the HIS3 coding sequence is replaced by that for GFP. The TetO construct replaces the poly(dA-dT) element from HIS3 with an operator for the tetracycline repressor protein (ACTCTATCAATGATAGAGT). The 154-bp spacer construct has 154 bp of nonyeast DNA between the GRE and LexAop.
FIG. 3.
Nonpositioned nucleosomes on the HIS3 upstream elements. (A) Primer extension analysis of nucleosome positioning using a primer upstream of the HIS3 start site. Naked DNA, DNA treated with MNase in vitro (MNase Naked), and chromatin prepared from the LexA− strain grown in glucose (Chm: Glu) were assayed. Similar results were obtained for chromatin obtained from the inducible LexA strain grown in galactose (data not shown). The schematic indicates the positions of the binding sites for LexA (black) and Gcn4 (white). (B) An indirect end-labeling assay of nucleosome positioning was performed with a probe downstream of HIS3 gene, DNSN, on samples from the analysis shown in panel A as well as on chromatin prepared from the inducible LexA strain grown in galactose (Chm: Gal). The gray box indicates the location of the LexA and Gcn4 binding sites.
FIG. 4.
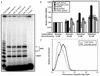
LexA coactivates gene expression in yeast. (A) Shown is an S1 assay for the inducible LexA strain grown in glucose (Glu) or galactose (Gal) with 0 or 10 mM aminotriazole (AT). The control sample is a parallel reaction lacking RNA. Total HIS3 mRNA levels from +1 and +13 initiation sites are quantified and normalized to DED1 levels. (B) The average HIS3 mRNA levels and standard errors from quantitative S1 assays are presented normalized to the glucose without aminotriazole condition for the following strains: LexA−, inducible LexA, mtGRE, mtLexAop, and 154-bp spacer. (C) Flow cytometric analysis was performed on GFP reporter strains grown in galactose. The histogram depicts the number of cells and their fluorescence intensity (log scale) for a representative run. The medians for LexA− and inducible LexA strains are 4.7 and 8.2 fluorescence units.
FIG. 5.
LexA increases occupancy at the neighboring Gcn4 binding site. Chromatin immunoprecipitation assays were performed with anti-Myc on inducible LexA strains containing either Myc epitope-tagged Gcn4 (Gcn4 Myc tag) or wild type Gcn4 (no tag). Strains were grown in glucose or galactose. Twenty or 40% of the whole-cell extract was used in the immunoprecipitation. PCR was performed with primers to the upstream element of our modified HIS3 construct (HIS3), of HIS4, and to a reference gene (Ref.), using total DNA (T) and precipitated DNA (20 and 40). A control reaction with no template DNA was also performed.
FIG. 6.
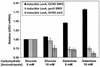
Gcn5 and Swi/Snf are required for HIS3 coactivation. Depicted are S1 assay results for the inducible LexA strain with wild-type Gcn5 and Swi/Snf (from the analysis shown in Fig. 4B), for the strain with Gcn5 deleted and for the strain with Swi2 deleted. Relative HIS3 mRNA levels were calculated as described in the legend for Fig. 4. Average values and standard errors are presented.
FIG. 7.
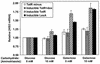
The tetracycline repressor coactivates gene expression in yeast. S1 assay results for the TetR− strain, the inducible TetR strain with or without doxycycline (dox), and the inducible LexA strain (from the analysis shown in Fig. 4B) are shown. Relative HIS3 mRNA levels were calculated as described in the legend for Fig. 4. Average values and standard errors are shown.
Similar articles
- The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5.
Sudarsanam P, Cao Y, Wu L, Laurent BC, Winston F. Sudarsanam P, et al. EMBO J. 1999 Jun 1;18(11):3101-6. doi: 10.1093/emboj/18.11.3101. EMBO J. 1999. PMID: 10357821 Free PMC article. - Activator control of nucleosome occupancy in activation and repression of transcription.
Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. Bryant GO, et al. PLoS Biol. 2008 Dec 23;6(12):2928-39. doi: 10.1371/journal.pbio.0060317. PLoS Biol. 2008. PMID: 19108605 Free PMC article. - Gcn4 occupancy of open reading frame regions results in the recruitment of chromatin-modifying complexes but not the mediator complex.
Topalidou I, Thireos G. Topalidou I, et al. EMBO Rep. 2003 Sep;4(9):872-6. doi: 10.1038/sj.embor.embor931. EMBO Rep. 2003. PMID: 12949586 Free PMC article. - Remodeling chromatin structures for transcription: what happens to the histones?
Steger DJ, Workman JL. Steger DJ, et al. Bioessays. 1996 Nov;18(11):875-84. doi: 10.1002/bies.950181106. Bioessays. 1996. PMID: 8939065 Review. - Affinity, Specificity, and Cooperativity of DNA Binding by Bacterial Gene Regulatory Proteins.
Carey J. Carey J. Int J Mol Sci. 2022 Jan 5;23(1):562. doi: 10.3390/ijms23010562. Int J Mol Sci. 2022. PMID: 35008987 Free PMC article. Review.
Cited by
- Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae.
Yarragudi A, Miyake T, Li R, Morse RH. Yarragudi A, et al. Mol Cell Biol. 2004 Oct;24(20):9152-64. doi: 10.1128/MCB.24.20.9152-9164.2004. Mol Cell Biol. 2004. PMID: 15456886 Free PMC article. - Identifying cooperativity among transcription factors controlling the cell cycle in yeast.
Banerjee N, Zhang MQ. Banerjee N, et al. Nucleic Acids Res. 2003 Dec 1;31(23):7024-31. doi: 10.1093/nar/gkg894. Nucleic Acids Res. 2003. PMID: 14627835 Free PMC article. - Sin mutations alter inherent nucleosome mobility.
Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Flaus A, et al. EMBO J. 2004 Jan 28;23(2):343-53. doi: 10.1038/sj.emboj.7600047. Epub 2004 Jan 15. EMBO J. 2004. PMID: 14726954 Free PMC article. - Chromatin remodelers clear nucleosomes from intrinsically unfavorable sites to establish nucleosome-depleted regions at promoters.
Tolkunov D, Zawadzki KA, Singer C, Elfving N, Morozov AV, Broach JR. Tolkunov D, et al. Mol Biol Cell. 2011 Jun 15;22(12):2106-18. doi: 10.1091/mbc.E10-10-0826. Epub 2011 Apr 20. Mol Biol Cell. 2011. PMID: 21508315 Free PMC article. - Structural constraints in collaborative competition of transcription factors against the nucleosome.
Moyle-Heyrman G, Tims HS, Widom J. Moyle-Heyrman G, et al. J Mol Biol. 2011 Sep 30;412(4):634-46. doi: 10.1016/j.jmb.2011.07.032. Epub 2011 Jul 29. J Mol Biol. 2011. PMID: 21821044 Free PMC article.
References
- Albrecht, G., H.-U. Mosch, B. Hoffmann, U. Reusser, and G. H. Braus. 1998. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:12696-12702. - PubMed
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases