Ends-out, or replacement, gene targeting in Drosophila - PubMed (original) (raw)
Ends-out, or replacement, gene targeting in Drosophila
Wei J Gong et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Ends-in and ends-out refer to the two arrangements of donor DNA that can be used for gene targeting. Both have been used for targeted mutagenesis, but require donors of differing design. Ends-out targeting is more frequently used in mice and yeast because it gives a straightforward route to replace or delete a target locus. Although ends-in targeting has been successful in Drosophila, an attempt at ends-out targeting failed. To test whether ends-out targeting could be used in Drosophila, we applied two strategies for ends-out gene replacement at the endogenous yellow (y) locus in Drosophila. First, a mutant allele was rescued by replacement with an 8-kb y(+) DNA fragment at a rate of approximately 1/800 gametes. Second, a wild-type gene was disrupted by the insertion of a marker gene in exon 1 at a rate of approximately 1/380 gametes. The I-SceI endonuclease component alone is not sufficient for targeting: the FLP recombinase is also needed to generate the extrachromosomal donor. When both components are used we find that ends-out targeting can be approximately as efficient as ends-in targeting, and is likely to be generally useful for Drosophila gene targeting.
Figures
Figure 1
Two general forms of gene targeting. Donor DNA molecules are diagramed above their targets, along with the expected products of recombination. The light gray region is the target-homologous DNA; the black region is the positive marker gene.
Figure 2
Constructs for ends-out targeting. Locations of y + and w + genes are indicated, along with the _FRT_s and the I-SceI recognition sequences (I-site). y u and y d indicate the upstream and downstream portions of y. The small arrowheads at the left and right ends of each construct indicate the P element inverted repeat termini.
Figure 3
Crossing schemes. Typical crosses used for the rescue and disruption experiments are shown. For the y rescue experiments, females with eye pigment were selected for the second cross, ensuring that they all carried the donor. For the disruption crosses, in some cases the donor element was hemizygous in the males, and so only half of the females used in the second cross carried the donor. The targeting frequency was adjusted accordingly in Table 1. In other cases we selected females exhibiting some degree of pigmentation in the eye, ensuring that they carried the donor; or the males used in the first cross carried the donor heterozygous with a dominantly marked balancer chromosome, thus allowing the selection of females that carried the donor for the second cross.
Figure 4
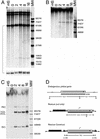
Verification of targeting. (A) Genomic Southern blotting to verify targeted y rescue. Letters above each lane indicate the genotype examined and refer to the expected structures of the donor construct, the y target gene (which is unchanged after the expected single-copy rescue event), and a tandem rescue event indicated to the right. The region used as a probe is indicated as a solid line below each structure, along with the expected sizes of the fragments produced by _Sph_I (Sp) digestion. The left and right lanes (M) carry molecular weight markers with sizes indicated at the left. The first four experimental lanes are targeted alleles; the next two are y w flies carrying the donor, and y w alone. (B) Cytological verification of targeted disruption. The w gene was used to probe polytene chromosomes of a larva carrying a targeted y allele. The two sites of hybridization, and their cytological designations, are indicated. (C) Genomic Southern blotting to verify targeted y disruption. The expected structures of the target locus, the donor construct, a targeted allele, and a tandem-insertion targeted allele are indicated to the right, along with the expected sizes of bands produced by Sal_I (S) digestion. The first two experimental lanes represent genomic DNA from y + w 1118 flies and from y+ w_1118 flies carrying the donor construct. The remainder are examples of targeting.
Figure 5
Efficiency of donor generation. Larvae carrying donor constructs and 70I-SceI (A and B) or 70FLP and 70I-SceI (C) were heat shocked at 38°C for 1 h. Genomic DNA was prepared at various times after heat shock (indicated in hours above the lanes). This experiment was carried out with a mixture of male and female larvae that were heterozygous for the donor element: 60% of the total yellow signal derives from the endogenous yellow gene; the remaining 40% comes from the donor. (A) Assay of DSBs at the I-SceI sites in the cut-only rescue construct (Fig. 2_C_). Genomic DNA was digested with Eco_RI. The 13.7-kb band (the largest band on the gel) and the 6.1-kb band are produced by the endogenous y gene. The band at ≈12 kb is one of the chromosomal donor bands generated by Eco_RI digestion (the other is not visible on this gel). The 6.3-/6.1-kb doublet is shown again at a reduced exposure at the bottom. (B) Genomic DNA from larvae carrying 70I-SceI and the cut-only construct, blotted without restriction digestion. The film was overexposed to visualize the 8.0-kb linear donor freed by I-SceI digestion. (C) Genomic DNA from larvae carrying 70FLP and 70I-SceI and the y rescue construct with FRTs (Fig. 2_A),_ blotted without restriction digestion. RC, relaxed circle; 1CL, single-cut linear; 2CL, double-cut linear; SC, supercoiled circle. (D) The structures of the endogenous y gene and the two donor constructs. All blots were probed with the 8-kb y probe indicated by the solid bar beneath the y gene. In the rescue construct, a single FRT adds ≈250 bp. R, _Eco_RI; -HS, not heat-shocked; MW, molecular weight markers.
Similar articles
- Methods for homologous recombination in Drosophila.
Maggert KA, Gong WJ, Golic KG. Maggert KA, et al. Methods Mol Biol. 2008;420:155-74. doi: 10.1007/978-1-59745-583-1_9. Methods Mol Biol. 2008. PMID: 18641946 - Targeted mutagenesis by homologous recombination in D. melanogaster.
Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Rong YS, et al. Genes Dev. 2002 Jun 15;16(12):1568-81. doi: 10.1101/gad.986602. Genes Dev. 2002. PMID: 12080094 Free PMC article. - Case studies of ends-out gene targeting in Drosophila.
Chen H, Ma Z, Liu Z, Tian Y, Xiang Y, Wang C, Scott MP, Huang X. Chen H, et al. Genesis. 2009 May;47(5):305-8. doi: 10.1002/dvg.20501. Genesis. 2009. PMID: 19298016 - Genome manipulation by homologous recombination in Drosophila.
Bi X, Rong YS. Bi X, et al. Brief Funct Genomic Proteomic. 2003 Jul;2(2):142-6. doi: 10.1093/bfgp/2.2.142. Brief Funct Genomic Proteomic. 2003. PMID: 15239936 Review. - Recombinases and their use in gene activation, gene inactivation, and transgenesis.
Bischof J, Basler K. Bischof J, et al. Methods Mol Biol. 2008;420:175-95. doi: 10.1007/978-1-59745-583-1_10. Methods Mol Biol. 2008. PMID: 18641947 Review.
Cited by
- The oscillating miRNA 959-964 cluster impacts Drosophila feeding time and other circadian outputs.
Vodala S, Pescatore S, Rodriguez J, Buescher M, Chen YW, Weng R, Cohen SM, Rosbash M. Vodala S, et al. Cell Metab. 2012 Nov 7;16(5):601-12. doi: 10.1016/j.cmet.2012.10.002. Epub 2012 Nov 1. Cell Metab. 2012. PMID: 23122660 Free PMC article. - The Basis of Food Texture Sensation in Drosophila.
Zhang YV, Aikin TJ, Li Z, Montell C. Zhang YV, et al. Neuron. 2016 Aug 17;91(4):863-877. doi: 10.1016/j.neuron.2016.07.013. Epub 2016 Jul 28. Neuron. 2016. PMID: 27478019 Free PMC article. - Combining recombineering and ends-out homologous recombination to systematically characterize Drosophila gene families: Rab GTPases as a case study.
Chan CC, Scoggin S, Hiesinger PR, Buszczak M. Chan CC, et al. Commun Integr Biol. 2012 Mar 1;5(2):179-83. doi: 10.4161/cib.18788. Commun Integr Biol. 2012. PMID: 22808327 Free PMC article. - The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization.
Bonnefoy E, Orsi GA, Couble P, Loppin B. Bonnefoy E, et al. PLoS Genet. 2007 Oct;3(10):1991-2006. doi: 10.1371/journal.pgen.0030182. Epub 2007 Sep 10. PLoS Genet. 2007. PMID: 17967064 Free PMC article. - Molecular evolution and functional characterization of Drosophila insulin-like peptides.
Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Grönke S, et al. PLoS Genet. 2010 Feb 26;6(2):e1000857. doi: 10.1371/journal.pgen.1000857. PLoS Genet. 2010. PMID: 20195512 Free PMC article.
References
- Rothstein R. Methods Enzymol. 1991;194:281–301. - PubMed
- Muller U. Mech Dev. 1999;82:3–21. - PubMed
- Rong Y S, Golic K G. Science. 2000;288:2013–2018. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases