Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II - PubMed (original) (raw)
Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II
Nicola Tolliday et al. Mol Biol Cell. 2003 Feb.
Abstract
In the budding yeast Saccharomyces cerevisiae, an actomyosin-based contractile ring is present during cytokinesis, as occurs in animal cells. However, the precise requirement for this structure during budding yeast cytokinesis has been controversial. Here we show that deletion of MYO1, the single myosin II gene, is lethal in a commonly used strain background. The terminal phenotype of myo1Delta is interconnected chains of cells, suggestive of a cytokinesis defect. To further investigate the role of Myo1p in cytokinesis, we conditionally disrupted Myo1 function by using either a dominant negative Myo1p construct or a strain where expression of Myo1p can be shut-off. Both ways of disruption of Myo1 function result in a failure in cytokinesis. Additionally, we show that a myo1Delta strain previously reported to grow nearly as well as the wild type contains a single genetic suppressor that alleviates the severe cytokinesis defects of myo1Delta. Using fluorescence time-lapse imaging and electron microscopy techniques, we show that cytokinesis in this strain is achieved through formation of multiple aberrant septa. Taken together, these results strongly suggest that the actomyosin ring is crucial for successful cytokinesis in budding yeast, but new cytokinetic mechanisms can evolve through genetic changes when myosin II function is impaired.
Figures
Figure 1
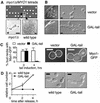
Myo1p is required for cytokinesis in the W303a background. (A) Phenotype of myo1Δ in W303a. Tetrad analysis of a diploid strain (RLY1236) heterozygous for the myo1Δ mutation showing 2:2 segregation for viability (upper panel). The plate was photographed after 2 d growth at 30°C. The morphology of a typical myo1Δ microcolony after 20 h growth at 30°C compared with a wild-type colony at the same stage (lower panels). Asterisk indicates that the myo1Δ microcolony did not increase further in size and eventually lysed. Note that the images are not to the same scale. (B) RLY1355 (vector control) and RLY884 (GAL-tail) were struck onto a YPGR plate and grown at 25°C for 3 d (left panels). A representative colony from each strain was picked for examination of cellular morphology (right panels). (C) RLY1451 (Myo1p-GFP plus vector control) and RLY1450 (Myo1p-GFP plus GAL-tail) cells were cultured overnight in YPR at 30°C. Galactose was added to induce tail expression, and samples were taken at 0 and 4 h. The number of cells in which Myo1p-GFP was localized to the bud neck was counted and divided by the total number of budded cells counted (at least 100) to give percent bud necks with Myo1p-GFP. Representative images showing Myo1p-GFP localization at the 4-h time point are shown next to the graph, with the cell outlines drawn in for clarity. (D) RLY261 (wild-type) and RLY884 (GAL-tail) cells were cultured overnight in YPR at 30°C. Galactose and α-factor (10 ng/ml final concentration) were added and the cells were grown at 30°C for 3 h. After washing three times with water, the cells were released into YPGR, and 5-ml duplicates were fixed with formaldehyde at 0, 2, and 4 h after release. Fixed cells were treated with zymolyase and counted on a hemacytometer. The resultant cell concentration at each time point was divided by that at time 0 to give relative cell number. Representative images of zymolyase-treated cells at the 4-h time point are also shown. Scale bars: A, 10 μm; B–D, 5 μm.
Figure 2
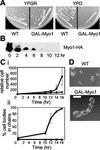
Cytokinesis defects caused by MYO1 shut-off. (A) Growth of wild-type (RLY261) and Myo1 shut-off (RLY1776) strains on YPGR or YPD plates, photographed after 3 d growth at 30°C. (B) RLY1776 cells were cultured overnight in YPGR at 30°C. A 10-ml sample was processed for immunoblot analysis (time 0). The rest of the cells were washed once with water and then shifted to YPD to shut off Myo1 expression. Samples were taken every 2 h after the shift and processed for immunoblotting using an anti-HA antibody to detect Myo1p level. (C) RLY261 (▪) and RLY1776 (●) strains were cultured and shifted as in B. (i) Samples were taken at 0, 10, 12, 14, and 16 h, fixed in formaldehyde, treated with zymolyase, and counted on a hemocytometer. Cell number calculation was done as described in Figure 1 legend. (ii) The percentage of cell bodies in chains was obtained by counting the number of cell bodies in chains of three or more cell bodies, divided by the total number of cell bodies in a defined field. The numbers at each time point in the plot were averages of duplicate samples. (D) Typical RLY 261 and RLY1776 cells from the 14-h time point are shown. Bar, 5 μm.
Figure 3
Phenotype of myo1Δ in BF264-Du background. (A) Tetrad analysis of a diploid strain (RLY1400) heterozygous for the myo1Δ mutation showing two distinct myo1Δ phenotypes, as represented by the boxed colonies (upper panel). The plate was photographed after 2 d growth at 30°C. An exponentially growing culture from each colony boxed above was fixed with formaldehyde and treated with zymolyase, and the cellular morphology was examined (lower panels). (B) Tetrad analysis of a diploid strain (RLY1468) created by mating two myo1Δ (healthy) strains, showing 100% robust growth. (C) Tetrad analysis of a diploid strain (RLY1488) created by mating myo1Δ (healthy) to myo1Δ (sick), showing 2:2 segregation for robust growth. Scale bar, 5 μm.
Figure 4
Chs2p dynamics in wild-type cells. (A) Chs2p-GFP, GFP-Tub1p expressing wild-type cells (RLY1673) were observed using three-dimensional (3D) confocal video microscopy (see online video material), as described in MATERIALS AND METHODS. A representative time-lapse series is shown, consisting of deconvolved 2D projections of each 3D image. Note that cell outlines have been drawn in for clarity. White arrows indicate signal from GFP-Tub1p, showing spindle disassembly. Scale bar, 3 μm. (B) Intensity profiles of bud neck localized Chs2p-GFP. A line was drawn through the Chs2p ring of the cell shown in A at each of the indicated time points, and pixel intensity values along the line were obtained, as described in MATERIALS AND METHODS. The measurements were then plotted against distance along the line.
Figure 5
Chs2p dynamics in myo1Δ (healthy) cells. (A and C) Chs2p-GFP, GFP-Tub1p expressing myo1Δ (healthy) cells (RLY1674) were observed using three-dimensional (3D) confocal video microscopy, as described in MATERIALS AND METHODS. Two representative time-lapse series are shown, consisting of deconvolved two-dimensional projections of each 3D image. Note that cell outlines have been drawn in for clarity. White arrows indicate bud neck localized Chs2p-GFP. Scale bar, 3 μm. (B and D) Intensity profiles of bud neck localized Chs2p-GFP. A line was drawn through the Chs2p ring of the cells shown in A and C, respectively, at each of the indicated time points, and pixel intensity values along the line were obtained, as described in MATERIALS AND METHODS. The measurements were then plotted against distance along the line.
Figure 6
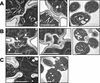
Aberrant septum formation in myo1Δ (healthy) cells. Wild-type (A), myo1Δ (healthy) (B), and myo1Δ (sick) (C) cells were cultured overnight and then arrested using 10 μg/ml nocodazole. Aliquots of cells were fixed at 30, 45, and 60 min after release from arrest and pooled (to enrich for cells undergoing cytokinesis), followed by processing for electron microscopic analysis. (A) Typical wild-type cells with only the primary (left panel) or both the primary and secondary (middle and right panels, two magnifications are shown) septa. (B) Typical myo1Δ (healthy) cells in the process of septum formation (left panel) or after septa formation (middle and right panels). White arrows indicate multiple membrane invaginations observed in myo1Δ (healthy). (C) Typical two budded myo1Δ (sick) cell that had failed cytokinesis and septum formation (two magnifications are shown). Scale bars, 1 μm.
Figure 7
A model to explain membrane closure events during cytokinesis in the wild-type or myo1Δ (healthy) cells. (A) In wild-type cells (left panel) contraction of the actomyosin ring drives ingression of the plasma membrane, pulling Chs2p in toward the center of the bud neck. Chitin is deposited (through the action of Chs2p) outside the membrane in the deepening cleavage furrow, and this continues until a disk of chitin separates the two cells (the primary septum). In myo1Δ (healthy) cells (right panel), the lack of an actomyosin ring and the presence of a suppressor mutation may result in Chs2p no longer being concentrated in one position at the bud neck, and multiple membrane invaginations are formed in various directions. Membrane closure in myo1Δ (healthy) cells occurs in a haphazard manner that results in the enclosure of large amounts of cytoplasm between multiple septa. (B) Selected images from time-lapse movies of Chs2p-GFP in wild-type (left) and myo1Δ (healthy) (right) were overlaid, as described in MATERIALS AND METHODS. In each case, the orientation of the Chs2p-GFP ring was compared with that of the mother-bud axis.
Similar articles
- Characterization of the minimum domain required for targeting budding yeast myosin II to the site of cell division.
Lister IM, Tolliday NJ, Li R. Lister IM, et al. BMC Biol. 2006 Jun 26;4:19. doi: 10.1186/1741-7007-4-19. BMC Biol. 2006. PMID: 16800887 Free PMC article. - Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast.
Lord M, Laves E, Pollard TD. Lord M, et al. Mol Biol Cell. 2005 Nov;16(11):5346-55. doi: 10.1091/mbc.e05-07-0601. Epub 2005 Sep 7. Mol Biol Cell. 2005. PMID: 16148042 Free PMC article. - Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis.
Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Bi E, et al. J Cell Biol. 1998 Sep 7;142(5):1301-12. doi: 10.1083/jcb.142.5.1301. J Cell Biol. 1998. PMID: 9732290 Free PMC article. - Actomyosin ring driven cytokinesis in budding yeast.
Meitinger F, Palani S. Meitinger F, et al. Semin Cell Dev Biol. 2016 May;53:19-27. doi: 10.1016/j.semcdb.2016.01.043. Epub 2016 Feb 1. Semin Cell Dev Biol. 2016. PMID: 26845196 Free PMC article. Review. - Mechanisms of cytokinesis in budding yeast.
Wloka C, Bi E. Wloka C, et al. Cytoskeleton (Hoboken). 2012 Oct;69(10):710-26. doi: 10.1002/cm.21046. Epub 2012 Jul 31. Cytoskeleton (Hoboken). 2012. PMID: 22736599 Review.
Cited by
- Characterization of the minimum domain required for targeting budding yeast myosin II to the site of cell division.
Lister IM, Tolliday NJ, Li R. Lister IM, et al. BMC Biol. 2006 Jun 26;4:19. doi: 10.1186/1741-7007-4-19. BMC Biol. 2006. PMID: 16800887 Free PMC article. - Transfer of the Septin Ring to Cytokinetic Remnants in ER Stress Directs Age-Sensitive Cell-Cycle Re-entry.
Chao JT, Piña F, Onishi M, Cohen Y, Lai YS, Schuldiner M, Niwa M. Chao JT, et al. Dev Cell. 2019 Oct 21;51(2):173-191.e5. doi: 10.1016/j.devcel.2019.08.017. Epub 2019 Sep 26. Dev Cell. 2019. PMID: 31564614 Free PMC article. - Regulation of contractile ring formation and septation in Schizosaccharomyces pombe.
Willet AH, McDonald NA, Gould KL. Willet AH, et al. Curr Opin Microbiol. 2015 Dec;28:46-52. doi: 10.1016/j.mib.2015.08.001. Epub 2015 Sep 3. Curr Opin Microbiol. 2015. PMID: 26340438 Free PMC article. Review. - Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis.
Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, Pringle JR. Ko N, et al. Mol Biol Cell. 2007 Dec;18(12):5139-53. doi: 10.1091/mbc.e07-05-0509. Epub 2007 Oct 17. Mol Biol Cell. 2007. PMID: 17942599 Free PMC article. - Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor.
Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Rancati G, et al. Cell. 2008 Nov 28;135(5):879-93. doi: 10.1016/j.cell.2008.09.039. Cell. 2008. PMID: 19041751 Free PMC article.
References
- Boyne JR, Yosuf HM, Bieganowski P, Brenner C, Price C. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J Cell Sci. 2000;113(Pt 24):4533–4543. - PubMed
- Cabib E, Roh DH, Schmidt M, Crottie LB, Varma A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J Biol Chem. 2001;276(23):19679–19682. - PubMed
- Cabib E, Ulane R, Bowers B. A molecular model for morphogenesis: the primary septum of yeast. Curr Top Cell Regul. 1974;8(0):1–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases