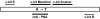One-step detection of c-kit point mutations using peptide nucleic acid-mediated polymerase chain reaction clamping and hybridization probes - PubMed (original) (raw)
One-step detection of c-kit point mutations using peptide nucleic acid-mediated polymerase chain reaction clamping and hybridization probes
Karl Sotlar et al. Am J Pathol. 2003 Mar.
Abstract
The prognostic significance of somatic activating codon 816 c-kit mutations in pediatric urticaria pigmentosa has not yet been established in detail. Detection of such mutations in archival paraffin-embedded biopsies is usually hampered by an abundance of surrounding normal cells. Here we describe a method for the selective amplification and specific detection of c-kit mutation Asp816-->Val in complete tissue sections cut from up to 24-year-old paraffin blocks. Peptide nucleic acid-mediated polymerase chain reaction clamping of the wild-type allele was combined with on-line mutation detection using oligonucleotide hybridization probes. In DNA extracted from HMC-1 cells heterozygously carrying the c-kit mutation Asp816-->Val, the one-tube assay allowed specific detection of this mutation in a more than 1000-fold excess of normal background DNA within 1 hour and without the need for additional analytical steps. In a series of 38 cases with pediatric urticaria pigmentosa we detected c-kit codons 815 and 816 mutations in 16 cases. Mutation detection did not correlate with clinical outcome after a mean follow-up of 11.2 years. In conclusion, the procedure described may represent an ideal screening tool for all kinds of clinical applications, using point mutations as markers of, for example, early events in carcinogenesis, circulating metastatic tumor cells, and minimal residual disease.
Figures
Figure 1.
Photomicrographs of pediatric UP showing dense subepidermal MC infiltrates without any preferential attachment to pre-existing structures. Molecular analysis disclosed wild-type c-kit in A and mutation Asp816→Val in B. Results of melting curve analyses are shown in Figure 4, A and B ▶ . Paraffin sections, AA1 immunostain; original magnifications, ×200.
Figure 2.
Positions of PCR primers (c-kit S, c-kit B), hybridization probes (c-kit mutation, c-kit anchor), and the PNA molecule targeting the wild-type sequence at codon 816 of the c-kit exon 17. Mutation Asp816→Val is caused by a A→T transition.
Figure 3.
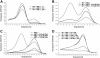
Effects of PNA-mediated PCR clamping. In melting curve analyses with Light Cycler hybridization probes wild-type (wt) c-kit and codon 17 mutation Asp816→Val (mut) can be discriminated through different melting peaks at ∼57°C (wt) and 63°C (mut) (A, continuous line). With only the use of hybridization probes, the method allows detection of the heterozygous mutation in an 10:1 excess of normal versus HMC-1 cells (A, broken line). Effects of PNA-mediated PCR clamping of the wild-type allele is demonstrated with various amounts of input PNA. In 100 ng of HMC-1 DNA, 0.75 μmol/L of PNA completely suppresses amplification of the wild-type allele (B, dotted line). Reduction of amplification efficiency of also the mutated allele by increasing amounts of PNA as a side effect must be taken into consideration when assessing the appropriate amount of input PNA (C). Compared to results in A, PNA-mediated PCR clamping of the wild-type allele results in a 100-fold increase of sensitivity yet allowing detection of the mutated allele in a 1000-fold excess of wild-type background DNA (D, three upper curves).
Figure 4.
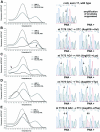
Results of melting curve analyses and direct sequencing of PCR products in five cases of pediatric UP (Table 1 ▶ ; patients 8, 9, 16, 19, and 22). In figures on the left melting curves of PCR products amplified without PNA (PNA−, dotted line) and with PNA-mediated PCR clamping (PNA+, broken line) are shown. As controls, melting curves of PNA-amplified HMC-1 DNA, heterozygous for c-kit mutation Asp816→Val, are included in every figure (continuous line). Melting curve analysis was performed with hybridization probes c-kit mutation and c-kit anchor. Figures on the right display corresponding results of direct sequencing of PNA− and PNA+ amplification products. In A no exon 17 mutation was observed. B represents 1 of 10 cases with an A→T transition in codon 816. G→A and G→T transversions of codons 815 and 816 are shown in C and D. A combination of the mutations in B and D is shown in E. This form of mutation involving codon 816 has been detected in two patients (Table 1 ▶ , patients 7 and 16).
Similar articles
- Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis.
Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Willmore-Payne C, et al. Hum Pathol. 2005 May;36(5):486-93. doi: 10.1016/j.humpath.2005.03.015. Hum Pathol. 2005. PMID: 15948115 - Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product.
Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfield JH, Ashman LK, Kanayama Y, et al. Furitsu T, et al. J Clin Invest. 1993 Oct;92(4):1736-44. doi: 10.1172/JCI116761. J Clin Invest. 1993. PMID: 7691885 Free PMC article. - Familial urticaria pigmentosa: report of a family and review of the role of KIT mutations.
Fett NM, Teng J, Longley BJ. Fett NM, et al. Am J Dermatopathol. 2013 Feb;35(1):113-6. doi: 10.1097/DAD.0b013e31826330bf. Am J Dermatopathol. 2013. PMID: 22892471 Review. - Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm.
Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, Duffy T, Jacobs P, Tang LH, Modlin I. Longley BJ, et al. Nat Genet. 1996 Mar;12(3):312-4. doi: 10.1038/ng0396-312. Nat Genet. 1996. PMID: 8589724 - K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization.
Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, Kensler TW, Bose KK, Cameron JL, Bos JL. Hruban RH, et al. Am J Pathol. 1993 Aug;143(2):545-54. Am J Pathol. 1993. PMID: 8342602 Free PMC article. Review.
Cited by
- Class III Receptor Tyrosine Kinases in Acute Leukemia - Biological Functions and Modern Laboratory Analysis.
Berenstein R. Berenstein R. Biomark Insights. 2015 Aug 5;10(Suppl 3):1-14. doi: 10.4137/BMI.S22433. eCollection 2015. Biomark Insights. 2015. PMID: 26309392 Free PMC article. Review. - Sensitive and reliable detection of KIT p.D816V mutation in decalcified archival bone marrow trephines.
Odensass M, Bartels S, Schlue J, Büsche G, Kreipe HH, Lehmann U. Odensass M, et al. Virchows Arch. 2025 Jun;486(6):1349-1353. doi: 10.1007/s00428-024-03973-8. Epub 2024 Nov 14. Virchows Arch. 2025. PMID: 39537990 Free PMC article. - Guidelines and diagnostic algorithm for patients with suspected systemic mastocytosis: a proposal of the Austrian competence network (AUCNM).
Valent P, Aberer E, Beham-Schmid C, Fellinger C, Fuchs W, Gleixner KV, Greul R, Hadzijusufovic E, Hoermann G, Sperr WR, Wimazal F, Wöhrl S, Zahel B, Pehamberger H. Valent P, et al. Am J Blood Res. 2013 May 5;3(2):174-80. Print 2013. Am J Blood Res. 2013. PMID: 23675567 Free PMC article. - Improved detection of the KIT D816V mutation in patients with systemic mastocytosis using a quantitative and highly sensitive real-time qPCR assay.
Kristensen T, Vestergaard H, Møller MB. Kristensen T, et al. J Mol Diagn. 2011 Mar;13(2):180-8. doi: 10.1016/j.jmoldx.2010.10.004. J Mol Diagn. 2011. PMID: 21354053 Free PMC article. - Germline mutations of KIT in gastrointestinal stromal tumor (GIST) and mastocytosis.
Ke H, Kazi JU, Zhao H, Sun J. Ke H, et al. Cell Biosci. 2016 Oct 18;6:55. doi: 10.1186/s13578-016-0120-8. eCollection 2016. Cell Biosci. 2016. PMID: 27777718 Free PMC article. Review.
References
- Valent P, Horny HP, Escribano L, Longley BJ, Li CY, Schwartz LB, Marone G, Nunez R, Akin C, Sotlar K, Sperr WR, Wolff K, Brunning RD, Parwaresch RM, Austen KF, Lennert K, Metcalfe DD, Vardiman JW, Bennett JM: Diagnostic criteria and classification of mastocytosis: a consensus proposal. Leuk Res 2001, 25:603-625 - PubMed
- Lennert K, Parwaresch MR: Mast cells and mast cell neoplasia: a review. Histopathology 1979, 3:349-365 - PubMed
- Horny HP, Parwaresch MR, Lennert K: Bone marrow findings in systemic mastocytosis. Hum Pathol 1985, 16:808-814 - PubMed
- Stein DH: Mastocytosis: a review. Pediatr Dermatol 1986, 3:365-375 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources