blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration - PubMed (original) (raw)
blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration
Kim D Finley et al. J Neurosci. 2003.
Abstract
A common feature of many human neurodegenerative diseases is the accumulation of insoluble ubiquitin-containing protein aggregates in the CNS. Although Drosophila has been helpful in understanding several human neurodegenerative disorders, a loss-of-function mutation has not been identified that leads to insoluble CNS protein aggregates. The study of Drosophila mutations may identify unique components that are associated with human degenerative diseases. The Drosophila blue cheese (bchs) gene defines such a novel degenerative pathway. bchs mutants have a reduced adult life span with the age-dependent formation of protein aggregates throughout the neuropil of the CNS. These inclusions contain insoluble ubiquitinated proteins and amyloid precursor-like protein. Progressive loss of CNS size and morphology along with extensive neuronal apoptosis occurs in aged bchs mutants. BCHS protein is widely expressed in the cytoplasm of CNS neurons and is present over the entire length of axonal projections. BCHS is nearly 3500 amino acids in size, with the last 1000 amino acids consisting of three functional protein motifs implicated in vesicle transport and protein processing. This region along with previously unidentified proteins encoded in the human, mouse, and nematode genomes shows striking homology along the full length of the BCHS protein. The high degree of conservation between Drosophila and human bchs suggests that study of the functional pathway of BCHS and associated mutant phenotype may provide useful insights into human neurodegenerative disorders.
Figures
Fig. 1.
The bchs region, mutations, protein structure, and species alignment. A, The location and extent of deletions uncovering bchs are illustrated. The map of the bchs genomic region and deletion boundaries is as described previously [Finley et al. (1998); see flybase website in Materials and Methods]. The location and arrangement of the_bchs_ and dsf genes are indicated, as are the 402Cy P-element (w+;dark gray) and P-elements generating_bchs_ mutations (bchs 3_and bchs4; light gray). The 402Cy P-element is the parent of the Df(2L)dsf3 and Df(2L)dsf4 deletions and the bchs3 insertion. It is carried on the CyO chromosome used as a wild-type control in aging experiments. B, The intron/exon structure of_bchs was determined by comparison and alignment of cDNA and genomic sequences. The locations of BEACH, WD40, and FYVE protein motifs and potential phosphorylation sites are indicated. Sites of P-element insertion are indicated. The extent of the_bchs5_ internal deletion (determined by DNA sequencing) is indicated by the hatched bar. A precise excision derivative, the_bchsrev1_ allele, was determined by DNA sequencing and is indicated by arrows. C,Drosophila, vertebrate, and nematode BCHS protein alignments were generated by comparison of the inferred amino acid sequences. The overall size, protein domains, and amino acid sequence of the different BCHS proteins are conserved, including the first 2500 amino acids possibly identifying functional motifs within this region. The identified domains within the C-terminal region have the highest amino acid identity over divergent phyla, reaching >80% over extended regions of the protein.
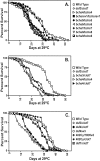
Fig. 2.
Premature death of bchs mutants. Newly eclosed males were collected and aged at 29°C for the duration of the experiments. A, Imprecise excisions of the_bchs_ 4 P-element, generating alterations in the 5′ region of the bchs gene, result in a shortened average life span, as shown by the_bchs5_/bchs5,bchs6/bchs6,bchs7/bchs7,bchs8/bchs8, and_bchs9_/bchs9_fly lines. Rescue of premature death occurs when the same_bchs4 P-element is removed precisely from its bchs location, as demonstrated by the_bchsrev1_/bchsrev1_line. B, P-element insertions in the first intron of_bchs reduce average life span, as seen in_bchs3_/Df(2L)clot7,bchs4/bchs4, and bchs4/Df(2L)clot7 genetic combinations. C, Flies containing one or more wild-type copies of bchs, Canton S, 402Cy/Df(2L)w3, or 402Cy/Df(2L)clot7 have an average life span between 29.0 and 32.5 d. Adults with both copies of the bchs genomic region removed, Df(2L)dsf3/Df(2L)clot7, Df(2L)dsf4/Df(2L)w3, or Df(2L)dsf4/Df(2L)clot7 flies, show a 40–45% reduction in average life span when compared with controls. Mutating the dsf gene in the case of dsf1/Df(2L)clot7 flies does not alter average adult longevity significantly. Mean lifespan, SDs, and p values for a selection of genotypes are summarized in Table 1.
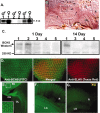
Fig. 3.
BCHS mRNA and protein expression patterns.A, Northern blot analysis of total RNA from 20 adult male and female heads. Wild-type (1) and_dsf_ 1/Df(2L)clot7 (3) mutant brains express a single, nearly 12.0 kb mRNA that is absent from Df(2L)dsf3/Df(2L)clot7 flies (2). B, Frozen sections from wild-type pharate adults (Finley et al., 1998) were used for in situ hybridization to bchs mRNA. Specific labeling for bchs is detected in a horizontal section through the head at the level of the antennal lobes. Labeling is seen in cortical regions containing neuronal cell bodies and is absent from neuropil areas. Arrows point to labeled cortical cells; the antenna lobe (AL), optic lobe (OL), and retina (R) are indicated. mRNA was not detected in muscles of the head (m) or other tissues. Sections through adult thorax also show uniform_bchs_ labeling in thoracic and abdominal ganglia neurons and not in other tissues (data not shown). C, Western blot analysis of BCHS shows a protein running well above the largest molecular weight marker (250 kDa) in head extracts from 1-d-old_Canton S_ (1) and 402CyO/Df(2L)clot7 (2) wild-type controls. At this age bchs4/Df(2L)clot7 flies (3) express some BCHS, whereas BCHS is absent from bchs3/Df(2L)clot7 (4) and Df(2L)dsf3/Df(2L)clot7 (5) flies. At 2 weeks only wild-type flies (1) have detectable levels of the BCHS protein. Protein no longer is detected in_bchs4_/Df(2L)clot7 flies (3). D–F, Subcellular location of the BCHS protein. D, Immunofluorescence staining of a third instar larval eye disc double-labeled for BCHS (basal section;FITC) and neuronal-specific marker ELAV (apical section;Texas Red). The two proteins demonstrate similar temporal expression patterns in neurons, but ELAV is located primarily within the nucleus while BCHS is found in cytoplasm (center; merged panel).E, Eye disc (ED), optic stalk (OS), and attached larval optic lobe (OL) are stained for BCHS (FITC). The protein is found along the entire length of photoreceptor axons. The inset is an enhanced intensity image showing BCHS extending as far as growth cones. This pattern is visible only because of the lack of BCHS expression in surrounding undifferentiated neuroblasts at this developmental time point. F, Mature BCHS expression pattern in a retina (R) and laminal ganglion (LG) optical section of a 2-week-old_dsf1_/Df(2L)clot7 adult (n = 9). BCHS protein levels are still high in older adults (consistent with Western blots). The protein remains located in the cytoplasm of retinal axons and in laminal ganglia.G, Enhanced image of an identically prepared retina and attached CNS from an age-matched bchs mutant [Df(2L)dsf3/Df(2L)clot7; n = 11]. This level of staining is similar to secondary antibody alone and is substantially below the staining of bchs+animals.
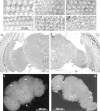
Fig. 4.
Neural degeneration and apoptosis in_bchs_ mutants. A–F, Plastic-embedded tangential sections (1 μm) of adult retina stained with methylene blue. Minor differences in early ommatidial morphology are attributable to the depth of each individual section and position within a given eye. A, Retinal morphology of Canton S_controls at 1 d of age is very similar to that of_bchs 3/Df(2L)clot7 (B) individuals of the same age.C, At 10 d wild-type controls demonstrate the mature architecture and cellular morphology of_Drosophila_ retina, whereas age-matched_bchs3_/Df(2L)clot7 flies (D) show the first signs of degeneration (vacuoles noted by arrows). E, At 14 d of age the wild-type retinas appear healthy and intact, with only the rare development of small vacuoles (arrow).F, At 14 d the degeneration detected earlier in_bchs3_/Df(2L)clot7 mutant flies has progressed further. There is a substantial loss of ommatidial architecture and an enlargement of vacuoles (arrows).G, H, Frontal sections (1 μm) from 10-d-old adults taken at the same depth within the head, near the central complex, as noted by the esophagus (E, arrows) and fan-shaped (FB) and ellipsoid bodies (EB). G, A section taken from 10-d-old controls demonstrates the characteristic organization and size of mature Drosophila neural structures. H, Age-matched section from a_bchs3_/Df(2L)clot7 fly shows the presence of retinal degeneration (arrow) as well as atrophy of laminal (LG) and medullary (MG) ganglia, inferior lateral deutocerebrum (ILD), superior lateral (SLPr) and medial (SMPr) protocerebrum, and subesophageal ganglia (SOG) structures of the CNS. Nuclei and background appear darker in this image because of slight image enhancement.I, J, In situ TUNEL analysis of 2-week-old brains. In wild-type brains (I) the arrows indicate the limited number of cells undergoing apoptosis [402CyO/Df(2L)clot7; n = 10]. bchs null brains (J) have extensive TUNEL labeling, indicating significant numbers of cells undergoing apoptosis in most cortical regions of the CNS [Df(2L)dsf3/Df(2L)clot7; n = 7].
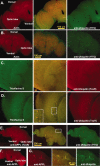
Fig. 5.
CNS protein aggregate formation.A–G, Single 0.2 μm transverse optical sections taken at least 5 μm into adult CNS. A, Optical section from an 11-d-old wild-type adult brain (n = 27) stained for actin (phalloidin-TRITC, Texas Red) and ubiquitin (FITC). Normal flies have a consistent pattern of actin and ubiquitin proteins within the CNS. Actin is enriched in neural projections and regions of synaptic innervation, whereas ubiquitin is stained uniformly throughout the entire CNS. B, Age-matched bchs null brains (n = 53) have deposits of ubiquitin in the CNS, mainly in regions of neuropil. C, Confocal images of 10-d-old wild-type controls (n = 20) stained for thioflavine S (green) and anti-ubiquitin (TexR).D, Identical images from age-matched_bchs_3/Df(2L)clot7 flies (n = 15) stained with thioflavine S and for ubiquitin aggregates. E, Confocal sections from 11-d-old Df(2L)dsf3/Df(2L)clot7 adult brain (n = 22) stained for APPL (TexR) and ubiquitin (FITC).Drosophila APPL colocalizes with ubiquitin deposits in the CNS. F, APPL aggregates do not form in 2-week-old wild-type controls (n = 12). G,Inset enlargement (through portions of the mushroom body) shows the close association of ubiquitin and APPL in protein aggregates.
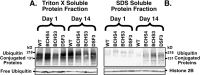
Fig. 6.
Immunoblot analysis of ubiquitin-conjugated proteins from adult Drosophila heads. Sequential protein extracts from heads of (1) wild-type, (2)bchs 4/Df(2L)clot7, (3)bchs3/Df(2L)clot7, and (4) Df(2L)dsf3/Df(2L)clot7 flies were examined by Western blot analysis for free ubiquitin and conjugated forms of the protein. A, In the Triton X-100 fraction (readily soluble proteins) the levels of free ubiquitin and high molecular weight ubiquitin-conjugated proteins were not altered significantly for any genotype or age that was examined (1 or 14 d). Significant differences in less soluble ubiquitin-conjugated proteins also were not detected in young animals after more stringent extractions (SDS fraction). B, At 14 d there is a substantial increase in the amount of less soluble high molecular weight ubiquitinated conjugates in the SDS fraction for all three bchs mutant combinations, but not for wild-type controls. The bchs4 allele (2) has a nearly ninefold increase, whereas the Df(2L)dsf3/Df(2L)clot7 deletion genotype (4) has the highest level of accumulation (22.8-fold increase) when compared with wild-type controls. Results are summarized in Table 1.
Similar articles
- Genetic modifiers of the Drosophila blue cheese gene link defects in lysosomal transport with decreased life span and altered ubiquitinated-protein profiles.
Simonsen A, Cumming RC, Lindmo K, Galaviz V, Cheng S, Rusten TE, Finley KD. Simonsen A, et al. Genetics. 2007 Jun;176(2):1283-97. doi: 10.1534/genetics.106.065011. Epub 2007 Apr 15. Genetics. 2007. PMID: 17435236 Free PMC article. - The Drosophila BEACH family protein, blue cheese, links lysosomal axon transport with motor neuron degeneration.
Lim A, Kraut R. Lim A, et al. J Neurosci. 2009 Jan 28;29(4):951-63. doi: 10.1523/JNEUROSCI.2582-08.2009. J Neurosci. 2009. PMID: 19176804 Free PMC article. - Linking lysosomal trafficking defects with changes in aging and stress response in Drosophila.
Simonsen A, Cumming RC, Finley KD. Simonsen A, et al. Autophagy. 2007 Sep-Oct;3(5):499-501. doi: 10.4161/auto.4604. Epub 2007 Jun 20. Autophagy. 2007. PMID: 17617737 - Swiss cheese et allii, some of the first neurodegenerative mutants isolated in Drosophila.
Kretzschmar D. Kretzschmar D. J Neurogenet. 2009;23(1-2):34-41. doi: 10.1080/01677060802471635. Epub 2009 Jan 7. J Neurogenet. 2009. PMID: 19132601 Free PMC article. Review. - Neurogenetics of courtship and mating in Drosophila.
Villella A, Hall JC. Villella A, et al. Adv Genet. 2008;62:67-184. doi: 10.1016/S0065-2660(08)00603-2. Adv Genet. 2008. PMID: 19010254 Review.
Cited by
- Characterization of axonal transport defects in Drosophila Huntingtin mutants.
Weiss KR, Littleton JT. Weiss KR, et al. J Neurogenet. 2016 Sep-Dec;30(3-4):212-221. doi: 10.1080/01677063.2016.1202950. Epub 2016 Jul 22. J Neurogenet. 2016. PMID: 27309588 Free PMC article. - ALFY localizes to early endosomes and cellular protrusions to facilitate directional cell migration.
Søreng K, Pankiv S, Bergsmark C, Haugsten EM, Dahl AK, de la Ballina LR, Yamamoto A, Lystad AH, Simonsen A. Søreng K, et al. J Cell Sci. 2022 Feb 15;135(4):jcs259138. doi: 10.1242/jcs.259138. Epub 2022 Feb 18. J Cell Sci. 2022. PMID: 35099014 Free PMC article. - Neurodegenerative mutants in Drosophila: a means to identify genes and mechanisms involved in human diseases?
Kretzschmar D. Kretzschmar D. Invert Neurosci. 2005 Nov;5(3-4):97-109. doi: 10.1007/s10158-005-0005-8. Epub 2005 Oct 24. Invert Neurosci. 2005. PMID: 16187075 Review. - Genetic modifiers of Drosophila palmitoyl-protein thioesterase 1-induced degeneration.
Buff H, Smith AC, Korey CA. Buff H, et al. Genetics. 2007 May;176(1):209-20. doi: 10.1534/genetics.106.067983. Epub 2007 Apr 3. Genetics. 2007. PMID: 17409080 Free PMC article. - Invertebrate models of neurologic disease: insights into pathogenesis and therapy.
Thompson LM, Marsh JL. Thompson LM, et al. Curr Neurol Neurosci Rep. 2003 Sep;3(5):442-8. doi: 10.1007/s11910-003-0028-7. Curr Neurol Neurosci Rep. 2003. PMID: 12914688 Review.
References
- Abrams JM. An emerging blueprint for apoptosis in Drosophila. Trends Cell Biol. 1999;9:435–440. - PubMed
- Alloway PG, Howard L, Dolph PJ. The formation of stable rhodopsin–arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron. 2000;28:129–138. - PubMed
- Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295:865–868. - PubMed
- Bacskai BJ, Kajdasz ST, Christie RH, Carter C, Games D, Seubert P, Schenk D, Hyman BT. Imaging of amyloid-β deposits in brains of living mice permits direct observation of clearance of plaques with immunotherapy. Nat Med. 2001;7:369–372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS09658/NS/NINDS NIH HHS/United States
- R01 NS009658/NS/NINDS NIH HHS/United States
- R01 GM56920A/GM/NIGMS NIH HHS/United States
- R03 AG19614/AG/NIA NIH HHS/United States
- R01 MH57460/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous