Protein ISGylation modulates the JAK-STAT signaling pathway - PubMed (original) (raw)
Protein ISGylation modulates the JAK-STAT signaling pathway
Oxana A Malakhova et al. Genes Dev. 2003.
Abstract
ISG15 is one of the most strongly induced genes upon viral infection, type I interferon (IFN) stimulation, and lipopolysaccharide (LPS) stimulation. Here we report that mice lacking UBP43, a protease that removes ISG15 from ISGylated proteins, are hypersensitive to type I IFN. Most importantly, in UBP43-deficient cells, IFN-beta induces a prolonged Stat1 tyrosine phosphorylation, DNA binding, and IFN-mediated gene activation. Furthermore, restoration of ISG15 conjugation in protein ISGylation-defective K562 cells increases IFN-stimulated promoter activity. These findings identify UBP43 as a novel negative regulator of IFN signaling and suggest the involvement of protein ISGylation in the regulation of the JAK-STAT pathway.
Figures
Figure 1
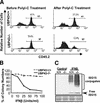
UBP43−/− mice are hypersensitive to the injection of a potent IFN inducer, polyI-C. (A) Seven mice of each group (UBP43+/+ and UBP43−/−) were intraperitoneally (ip) injected daily with polyI-C. Viability was monitored daily. (B) UBP43+/+ and UBP43−/− mice (n = 4 for each group) were ip-injected daily with polyI-C. The total number of white blood cells (WBCs) in the peripheral blood was counted daily after each polyI-C injection. (C) UBP43+/+ and UBP43−/− mice (n = 5 for each group) were either left untreated or ip-injected with polyI-C 24 h prior to bone marrow extraction. Bone marrow cells were counted in the control and polyI-C-injected groups of mice (two femurs from each mouse).
Figure 2
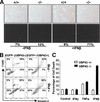
The hypersensitivity to IFN stimulation is intrinsic in UBP43-deficient hematopoietic cells. (A) Hematopoietic cells transplanted from UBP43−/− mice to UBP43+/+ congenic strain mice are hypersensitive to polyI-C injection. Bone marrow cells (CD45.2+) from individual UBP43+/+ and UBP43−/− mice (n = 4 for each group) were isolated and transplanted to lethally irradiated congenic strain mice (CD45.2−). Six months after the transplantation, flow cytometric analysis was done to determine the relative amount of peripheral blood cells contributed from the donor or recipient mice based on CD45.2 immunostaining before and after polyI-C injection (48 h). Flow cytometric analysis results from one representative mouse transplanted with UBP43+/+ or UBP43−/− bone marrow are presented. (B) UBP43−/− bone marrow cells are hypersensitive to IFN-β treatment. Bone marrow cells from UBP43+/+ and UBP43−/− mice were cultured at 2 × 104 cells/mL in CFU assay medium in the presence of various concentrations of IFN-β. Total colony number was counted after 12 d of culture. Results of a single representative experiment of two independent experiments are shown. Data points represent mean (±S.D.) colony growth derived from colony counts in three replicate plates for each mouse with two mice in each group. (C) Increased level of ISG15 conjugation in UBP43−/− bone marrow cells. Bone marrow cells were either left untreated or stimulated with 100 unit/mL IFN-β for 24 h. Total proteins (15 μg) were resolved on 6%–18% gradient SDS-PAGE and immunoblotted with anti-mouse ISG15 antibodies (Abs). The bottom panel shows the membrane stained by Ponceau S.
Figure 3
Augmented apoptosis rate of UBP43−/− cells is specific to type I IFN treatment. (A) Bone marrow cells isolated from UBP43+/+ and UBP43−/− mice were cultured in vitro in the presence or the absence of INF-β (100 units/mL) for 48 h. A TUNEL assay was performed to study the percentage of apoptosis cells. (B) Forty-eight hours after UBP43−/− bone marrow cells were infected with UBP43-MigR1, they were cultured in the presence or absence of INF-β (100 units/mL) for 48 h. The cells (15% EGFP+ and 85% EGFP−) were stained with Annexin V-PE and 7-AAD and analyzed by flow cytometry. (C) Bone marrow cells from UBP43+/− and UBP43−/− mice were cultured in the absence or presence of 10 ng/mL INF-γ, 10 ng/mL TNF-α, and 100 units/mL INF-β for 48 h. Cells were then subjected to a Trypan blue dye exclusion assay. Each data point represents the mean of three independent experiments. Error bars represent standard deviation of means.
Figure 4
JAK-STAT pathway is extensively activated in UBP43−/− cells upon IFN stimulation. (A) ISGF3 complex formation in UBP43-deficient cells upon IFN-β stimulation. UBP43+/+ and UBP43−/− bone marrow cells were either left untreated or stimulated for the time indicated. Total protein extracts (30 μg) were used in the gel shift assays with 32P-labeled double-stranded oligonucleotide that contains an ISGF3 binding site. (B) UBP43+/+ and UBP43−/− bone marrow cells were either left untreated or stimulated with 100 unit/mL IFN-β as described above. Total proteins (15 μg) were resolved on 7% SDS-PAGE and immunoblotted with anti-phospho-Stat1 (Tyr701) Abs. Blots were stripped and reprobed with anti-Stat1 Abs to assure equal loading. (C) Induction of ISGs in UBP43+/+ and UBP43−/− bone marrow cells. Northern blot analysis of ISG15, 2‘-5′OAS and IRF7 gene expression was performed on total RNA isolated from UBP43+/+ and UBP43−/− bone marrow cells treated with 100 unit/mL of IFN-β for the time indicated.
Figure 5
Protein ISGylation increases IFN signaling. (A) functional UBE1L expression restores protein ISGylation in K562 cells. Control or UBE1L expression plasmid pcDNA-HA-UBE1L transiently transfected K562 cells were cultured in the absence or presence of 1000 units/mL IFN-α for 24 h. Protein extracts were prepared from these cells and used in the Western blot analysis with antibodies against ISG15 (top panel) and HA tag (bottom panel). Clear UBE1L expression can be detected in transfected cells. Asterisks mark several nonspecific signal bands. Positions of molecular weight markers are shown on the left side. (B) K562 cells were transiently transfected with p3K-UBP43-luc and either vector pcDNA or the UBE1L expression construct pcDNA-HA-UBE1L with Renilla luciferase expression construct pRL-null for transfection efficiency control. Cells were cultured in the presence or absence of 1000 units/mL IFN-α for the indicated time and harvested for a dual luciferase assay. The promoter activity is presented as relative firefly luciferase activity, which has been normalized to Renilla luciferase activity. The average promoter activity was generated from four separate experiments. Error bars represent standard deviation of the mean.
Similar articles
- UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity.
Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. Malakhova OA, et al. EMBO J. 2006 Jun 7;25(11):2358-67. doi: 10.1038/sj.emboj.7601149. Epub 2006 May 18. EMBO J. 2006. PMID: 16710296 Free PMC article. - Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling.
Kim KI, Malakhova OA, Hoebe K, Yan M, Beutler B, Zhang DE. Kim KI, et al. J Immunol. 2005 Jul 15;175(2):847-54. doi: 10.4049/jimmunol.175.2.847. J Immunol. 2005. PMID: 16002682 - Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation.
Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Kim KI, et al. Mol Cell Biol. 2004 Nov;24(21):9592-600. doi: 10.1128/MCB.24.21.9592-9600.2004. Mol Cell Biol. 2004. PMID: 15485925 Free PMC article. - [Molecular mechanisms for suppression of interferon signal transduction pathways caused by viral infections].
Fujii N, Yokota S, Yokosawa N, Okabayashi T. Fujii N, et al. Uirusu. 2004 Dec;54(2):169-78. doi: 10.2222/jsv.54.169. Uirusu. 2004. PMID: 15745154 Review. Japanese. - IL-3 signaling and the role of Src kinases, JAKs and STATs: a covert liaison unveiled.
Reddy EP, Korapati A, Chaturvedi P, Rane S. Reddy EP, et al. Oncogene. 2000 May 15;19(21):2532-47. doi: 10.1038/sj.onc.1203594. Oncogene. 2000. PMID: 10851052 Review.
Cited by
- Type I interferon regulation by USP18 is a key vulnerability in cancer.
Jové V, Wheeler H, Lee CW, Healy DR, Levine K, Ralph EC, Yamaguchi M, Jiang ZK, Cabral E, Xu Y, Stock J, Yang B, Giddabasappa A, Loria P, Casimiro-Garcia A, Kessler BM, Pinto-Fernández A, Frattini V, Wes PD, Wang F. Jové V, et al. iScience. 2024 Mar 27;27(4):109593. doi: 10.1016/j.isci.2024.109593. eCollection 2024 Apr 19. iScience. 2024. PMID: 38632987 Free PMC article. - Bulk and single cells transcriptomes with experimental validation identify USP18 as a novel glioma prognosis and proliferation indicator.
Chen Y, Li R, Li Z, Yang B, He J, Li J, Li P, Zhou Z, Wu Y, Zhao Y, Guo G. Chen Y, et al. Exp Ther Med. 2024 Mar 26;27(5):229. doi: 10.3892/etm.2024.12517. eCollection 2024 May. Exp Ther Med. 2024. PMID: 38596661 Free PMC article. - In the moonlight: non-catalytic functions of ubiquitin and ubiquitin-like proteases.
Campos Alonso M, Knobeloch KP. Campos Alonso M, et al. Front Mol Biosci. 2024 Feb 22;11:1349509. doi: 10.3389/fmolb.2024.1349509. eCollection 2024. Front Mol Biosci. 2024. PMID: 38455765 Free PMC article. Review. - Reprogramming of tumor-associated macrophages via NEDD4-mediated CSF1R degradation by targeting USP18.
Miyauchi S, Arimoto KI, Liu M, Zhang Y, Zhang DE. Miyauchi S, et al. Cell Rep. 2023 Dec 26;42(12):113560. doi: 10.1016/j.celrep.2023.113560. Epub 2023 Dec 13. Cell Rep. 2023. PMID: 38100351 Free PMC article. - ISGylation-independent protection of cell growth by USP18 following interferon stimulation.
Clancy A, Rusilowicz-Jones EV, Wallace I, Swatek KN, Urbé S, Clague MJ. Clancy A, et al. Biochem J. 2023 Oct 11;480(19):1571-1581. doi: 10.1042/BCJ20230301. Biochem J. 2023. PMID: 37756534 Free PMC article.
References
- Blomstrom DC, Fahey D, Kutny R, Korant BD, Knight E., Jr Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J Biol Chem. 1986;261:8811–8816. - PubMed
- Broxmeyer HE, Lu L, Platzer E, Feit C, Juliano L, Rubin BY. Comparative analysis of the influences of human γ, α and β interferons on human multipotential (CFU-GEMM), erythroid (BFU-E), and granulocyte-macrophage (CFU-GM) progenitor cells. J Immunol. 1983;131:1300–1305. - PubMed
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous