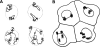Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages - PubMed (original) (raw)
Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages
Joachim Walter et al. J Cell Biol. 2003.
Abstract
Whether chromosomes maintain their nuclear positions during interphase and from one cell cycle to the next has been controversially discussed. To address this question, we performed long-term live-cell studies using a HeLa cell line with GFP-tagged chromatin. Positional changes of the intensity gravity centers of fluorescently labeled chromosome territories (CTs) on the order of several microm were observed in early G1, suggesting a role of CT mobility in establishing interphase nuclear architecture. Thereafter, the positions were highly constrained within a range of approximately 1 microm until the end of G2. To analyze possible changes of chromosome arrangements from one cell cycle to the next, nuclei were photobleached in G2 maintaining a contiguous zone of unbleached chromatin at one nuclear pole. This zone was stably preserved until the onset of prophase, whereas the contiguity of unbleached chromosome segments was lost to a variable extent, when the metaphase plate was formed. Accordingly, chromatin patterns observed in daughter nuclei differed significantly from the mother cell nucleus. We conclude that CT arrangements were stably maintained from mid G1 to late G2/early prophase, whereas major changes of CT neighborhoods occurred from one cell cycle to the next. The variability of CT neighborhoods during clonal growth was further confirmed by chromosome painting experiments.
Figures
Figure 1.
Chromosome arrangements in blastomere nuclei of P. equorum (2 n = 2) drawn by Theodor Boveri. (A) The two nuclei above and below each represent a pair of daughter nuclei from blastomeres studied at prophase of the two-cell stage. Chromosome ends are fixed within evaginations of the nuclear envelope. Note that chromosome arrangements and the positions of the evaginations are similar in each pair, whereas different pairs show striking differences. (B) Interphase blastomere cells from an embryo drawn at the four-cell stage. Chromosome arrangements within the nucleus are invisible, except for nuclear evaginations that indicate telomere positions. Each pair of daughter nuclei shows symmetrical positions of the evaginations, whereas a comparison of the two pairs reveals gross differences.
Figure 2.
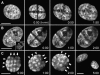
Stability of large-scale CT arrangements during interphase of HeLa cells studied by nuclear stripe photobleaching experiments. Cross-stripe (rows A and B) or mesh-like (row C) geometrical patterns bleached into HeLa cell nuclei with GFP-tagged H2B at different stages of the cell cycle. The patterns were maintained until recovery of GFP-fluorescence (1–2 h). (A) Nucleus bleached in G1 (2 h and 20 min after prophase of the mother nucleus). (B) S-phase nucleus. The nucleus was labeled with Cy5-dUTP before bleaching by scratch-replication labeling of the cell, and shows a labeling pattern typical for early S-phase (inset). Stripe photobleaching was performed 2 h later, ensuring that the cell was in middle S-phase at that time. (C) Late S/G2 nucleus. The cell entered mitosis 3 h after stripe bleaching. Note that the mesh-like pattern was stably maintained (arrows), whereas the nucleus rotates. All images present best focus light-optical sections. Bars, 10 μm.
Figure 3.
Stability of large-scale CT arrangements studied in nuclei with Cy3-labeled CTs. (A–D) Confocal time-lapse series of a HeLa cell (Table I, nucleus 3_m_) with replication-labeled CTs and its daughters recorded for a total observation period of 18 h. CT movements in the mother cell nucleus were analyzed from late G1 to late G2 (13-h evaluation period). Time point 0:00 indicates the start of evaluation. (A) Superimposed maximum intensity projections of confocal serial sections from H2B-GFP (green) and Cy3-dUTP (red) together with a transmission image at selected time points (arrowheads in A point to nuclei reconstructed in B). The start of evaluation was assigned to late G1 as the cell entered prophase 13.5 h later. (B) Top and side view of 3-D reconstructions for time points 0:00 (h:min), 4:30, 8:30, and 12:30. Nuclei (outlined in gray) are displayed after correction for rotational movements, and nuclear volumes are indicated. Four signal clusters representing different CTs are shown in different colors for easier discrimination. (C and D) Absolute and corrected distances between the fluorescence gravity center of each CT and the CN (CT-CN), as well as distances between all pairs of CTs (CT-CT) were evaluated at intervals of 30 min. Δ(max-min) indicates the difference between the maximum and minimum value of the respective distances within the complete evaluation period. (E–H) Confocal time-lapse series from another HeLa cell (Table I, nucleus 4) recorded with a time interval of 6 min. The 8-h evaluation period started at telophase (time point 0:00) and ended likely at early S-phase (8:00). The mother cell performed mitosis at time point −0:30. (E) Maximum intensity projections of confocal serial sections as described in A for selected time points (arrowheads in E point to nuclei reconstructed in F). (F) Top and side view of 3-D reconstructions from the evaluated nucleus containing 3 CTs. (G and H) Absolute and corrected distances as described above for C and D. CT-CN and CT-CT distances were evaluated at intervals of 12 min (early G1) and 30 min (mid G1–early S). The transition from early G1 to mid G1 is defined as the time point when the nuclear volume increase reaches a first plateau (see Materials and methods). Note that the higher fluctuation of distances in early G1 as compared with other cell cycle stages. Arrowheads in A and E indicate nuclei shown as 3-D reconstructions in B and F. Bars, 10 μm.
Figure 4.
Stability of CT neighborhood during interphase of living HeLa cells. HeLa cells were replication-labeled during S-phase of two consecutive cell cycles (first cycle, Cy3-dUTP, false color red; second cycle, Cy5-dUTP, false color green). Frames of maximum intensity projections from light optical serial sections are displayed for the indicated time points. Insets show GFP signals of confocal nuclear midsections in gray. Images are corrected for translational and rotational nuclear movements. Observation started 4.5 d after the second labeling event. After 3h 50 min, the cell entered prophase. Note that chromosome condensation is a locally confined process with little changes of chromosome arrangements (compare C with D). After completion of mitosis, daughter nuclei were followed for another 3.5 h. Locally constrained movements, but no major rearrangements of differently colored chromatin domains were detected (see Video 2). Bar, 5 μm.
Figure 5.
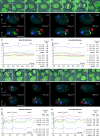
Large-scale CT arrangements in HeLa cell nuclei change from one cell cycle to the next. A–C show examples of live-cell confocal image series from three HeLa cells (for movie sequences, see Video 3). After bleaching of GFP-labeled chromatin, except for a contiguous unbleached region at one nuclear pole, cells were followed through the remaining part of interphase and through mitosis until the formation of daughter nuclei. All images are maximum intensity projections of confocal image stacks. The degree of the redistribution of unbleached chromatin patches in G1 daughter cells (column VI, arrowheads) can vary from “clustered” (A) to “partially clustered” (B) to “scattered” (C). Column I, GFP pattern of HeLa nuclei before bleaching; column II, the same nuclei after partial bleaching; column III, last frame recorded before the formation of the metaphase plate; column IV, metaphase plate. Arrowheads in B and C point to unbleached chromosomal fragments that are remote from the bulk of unbleached chromatin. Columns V and VI, daughter nuclei in anaphase (column V, C), telophase, or very early G1 (column V, A and B) and in early G1 (column VI). Bars, 10 μm.
Figure 6.
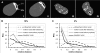
RAC of the distribution of unbleached chromatin in mother and daughter cell nuclei. (A) Projections of the mid-nuclear optical sections from a mother nucleus immediately after partial bleaching (I) and shortly before the onset of prophase (II), as well as from its two daughters in early G1 (III and IV; compare with Figure 5 C). Nuclear area shown in dark gray. Pixels of the 2% highest intensity fraction shown in black color belong exclusively to the unbleached zone of the mother nucleus, but do not completely cover it. Pixels of the 10% highest intensity fraction shown in white color fully cover the unbleached zone, but include a few pixels from chromatin in the bleached nuclear area (B and C). Average RAC curves calculated from the 10 and 2% highest intensity fraction for 24 mother cell nuclei immediately after bleaching and before prophase, as well as for the 48 daughter cell nuclei. For comparison, the RAC curve for mother nuclei immediately before bleaching is shown, taking into account all nuclear pixels. SDs are within the same order for mother and daughter cell nuclei, and are only shown for the latter.
Figure 7.
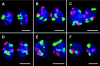
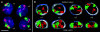
CT #7 and #10 arrangements in nuclei of two-cell clones. Projections of confocal image stacks obtained after painting of chromosome #10 (visualized in red) and #7 (visualized in green). DNA counterstain, blue. A, B, and D represent daughter nuclei with an obvious (although not perfect) symmetry of the relative CT arrangements. F provides an example for a large deviation from a symmetrical arrangement. To facilitate assessment of symmetry, some of the nuclei were rotated and/or mirrored. Bars, 10 μm.
Figure 8.
CT #7 and #10 arrangements in a four-cell clone. (A) Projection of a confocal image stack through nuclei n1 to n4 after painting of chromosome #10 (visualized in red) and #7 (visualized in green). DNA counterstain, blue. (B) 3-D reconstructions of n1 to n4 by volume rendering. Part of the nuclear border is indicated by reconstruction of the counterstain periphery (outside, blue; inside, silver-gray). The top row shows the bottom view, the bottom row the corresponding side view. To show the symmetry CT arrangements in nuclei n1 and n2 more clearly, the x-axis of n2 was inverted. Bar, 10 μm. For interactive videos of nuclear reconstructions, see supplemental information.
Comment in
- An uncertainty principle in chromosome positioning.
Parada LA, Roix JJ, Misteli T. Parada LA, et al. Trends Cell Biol. 2003 Aug;13(8):393-6. doi: 10.1016/s0962-8924(03)00149-1. Trends Cell Biol. 2003. PMID: 12888289
Similar articles
- Chromosomes, positions please!
Williams RR, Fisher AG. Williams RR, et al. Nat Cell Biol. 2003 May;5(5):388-90. doi: 10.1038/ncb0503-388. Nat Cell Biol. 2003. PMID: 12724773 Review. No abstract available. - Arrangement of nuclear structures is not transmitted through mitosis but is identical in sister cells.
Orlova DY, Stixová L, Kozubek S, Gierman HJ, Šustáčková G, Chernyshev AV, Medvedev RN, Legartová S, Versteeg R, Matula P, Stoklasa R, Bártová E. Orlova DY, et al. J Cell Biochem. 2012 Nov;113(11):3313-29. doi: 10.1002/jcb.24208. J Cell Biochem. 2012. PMID: 22644811 - Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture.
Cremer T, Kreth G, Koester H, Fink RH, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Cremer T, et al. Crit Rev Eukaryot Gene Expr. 2000;10(2):179-212. Crit Rev Eukaryot Gene Expr. 2000. PMID: 11186332 Review.
Cited by
- Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression.
Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ. Malhas A, et al. J Cell Biol. 2007 Feb 26;176(5):593-603. doi: 10.1083/jcb.200607054. Epub 2007 Feb 20. J Cell Biol. 2007. PMID: 17312019 Free PMC article. - Activation of estrogen-responsive genes does not require their nuclear co-localization.
Kocanova S, Kerr EA, Rafique S, Boyle S, Katz E, Caze-Subra S, Bickmore WA, Bystricky K. Kocanova S, et al. PLoS Genet. 2010 Apr 22;6(4):e1000922. doi: 10.1371/journal.pgen.1000922. PLoS Genet. 2010. PMID: 20421946 Free PMC article. - Maintenance of imprinting and nuclear architecture in cycling cells.
Teller K, Solovei I, Buiting K, Horsthemke B, Cremer T. Teller K, et al. Proc Natl Acad Sci U S A. 2007 Sep 18;104(38):14970-5. doi: 10.1073/pnas.0704285104. Epub 2007 Sep 11. Proc Natl Acad Sci U S A. 2007. PMID: 17848516 Free PMC article. - Gene positioning.
Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A. Ferrai C, et al. Cold Spring Harb Perspect Biol. 2010 Jun;2(6):a000588. doi: 10.1101/cshperspect.a000588. Epub 2010 May 19. Cold Spring Harb Perspect Biol. 2010. PMID: 20484389 Free PMC article. Review. - Blank spots on the map: some current questions on nuclear organization and genome architecture.
Adriaens C, Serebryannyy LA, Feric M, Schibler A, Meaburn KJ, Kubben N, Trzaskoma P, Shachar S, Vidak S, Finn EH, Sood V, Pegoraro G, Misteli T. Adriaens C, et al. Histochem Cell Biol. 2018 Dec;150(6):579-592. doi: 10.1007/s00418-018-1726-1. Epub 2018 Sep 20. Histochem Cell Biol. 2018. PMID: 30238154 Free PMC article. Review.
References
- Barlow, R. 1989. Statistics. Wiley, Chichester, UK. 204 pp.
- Baxter, J., M. Merkenschlager, and A.G. Fisher. 2002. Nuclear organisation and gene expression. Curr. Opin. Cell Biol. 14:372–376. - PubMed
- Bland, M. 2000. An Introduction to Medical Statistics. Oxford University Press, Oxford, UK. 424 pp.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials