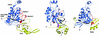Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 - PubMed (original) (raw)
Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Sarah M Garrard et al. EMBO J. 2003.
Abstract
Cdc42 is a small GTPase that is required for cell polarity establishment in eukaryotes as diverse as budding yeast and mammals. Par6 is also implicated in metazoan cell polarity establishment and asymmetric cell divisions. Cdc42.GTP interacts with proteins that contain a conserved sequence called a CRIB motif. Uniquely, Par6 possesses a semi-CRIB motif that is not sufficient for binding to Cdc42. An adjacent PDZ domain is also necessary and is required for biological effects of Par6. Here we report the crystal structure of a complex between Cdc42 and the Par6 GTPase-binding domain. The semi-CRIB motif forms a beta-strand that inserts between the four strands of Cdc42 and the three strands of the PDZ domain to form a continuous eight-stranded sheet. Cdc42 induces a conformational change in Par6, detectable by fluorescence resonance energy transfer spectroscopy. Nuclear magnetic resonance studies indicate that the semi-CRIB motif of Par6 is at least partially structured by the PDZ domain. The structure highlights a novel role for a PDZ domain as a structural scaffold.
Figures
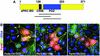
Fig. 1. (A) Schematic of Par6B domain structure. (B) Inhibition of tight junction assembly by the GBD of Par6B. MDCK canine epithelial cells were transiently transfected with vectors that express myc-tagged versions of the Par6B GBD (semi-CRIB + PDZ; residues 126–253), the isolated PDZ domain (140–256) or the semi-CRIB motif alone (102–186). The cells were incubated in a calcium-free medium overnight to disrupt cell–cell contacts, then returned to normal DMEM/serum for 6 h to permit junction re-assembly. They were then washed and stained for the tight junction marker, Z0-1 (green), the myc tag (red) and DNA (blue).
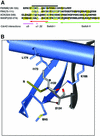
Fig. 2. Crystal structure of Par6B(126–253) bound to Cdc42(Q61L)·GMPPNP at 2.1 Å resolution. Ribbon diagram with Cdc42 colored blue and Par6 yellow (left and center images). A structure of the WASP–Cdc42 complex (Abdul-Manan et al., 1999) is also shown on the right, for comparison. The Switch I and II regions of Cdc42 are colored red, the semi-CRIB motif of Par6 and the CRIB motif of WASP are colored black, and the GMP-PNP/Mg2+(H2O)2 moiety is presented in ball-and-stick representation. A short region of the PDZ domain of Par6 (residues 204–208) that makes contact with Cdc42 is highlighted in magenta. The center image is of the complex rotated by 90° about the vertical axis. The black arrow indicates the location of the PDZ ligand-binding pocket.
Fig. 3. (A) Alignment of Par6B semi-CRIB motif with CRIB sequences of PAK, ACK and WASP, and Cdc42 residues involved in binding Par6B. The black line indicates the extent of the consensus CRIB motif. Conserved residues are shown in blue; residues that interact with Cdc42 are boxed in yellow, with the corresponding region of Cdc42 marked beneath. Cdc42 residues are colored to indicate structural regions. (B) Sequence-specific interactions of the N-terminal portion of the semi-CRIB motif of Par6 with Cdc42. The coloring scheme is similar to Figure 2, with the semi-CRIB motif of Par6 colored in light gray for clarity. The labels for the residues of the consensus CRIB sequence motif are italicized. Dotted black lines represent salt bridges and hydrogen bonds. Non-sequence-specific hydrogen bonds (e.g., the main chain hydrogen bonds of the β-sheet) are not shown for clarity.
Fig. 4. (A) Structure of the central region of a representative conformer of the CRIB motif of WASP with Cdc42 (PDB ID code 1cee). The coloring scheme is as shown in Figure 3. (B) For comparison, the sequence-specific interactions of the semi-CRIB motif of Par6 are shown in a similar orientation as in (A). (C) Asp38 is not required for binding of Par6 to Cdc42. Myc-tagged Par6B or, as a control, the CRIB domain of α-PAK, was immunoprecipitated from cells co-expressing GST fusions of Cdc42(Val12) that contain second mutations in the Switch I region. Bound GST–Cdc42 was detected by blotting with anti-GST antibody.
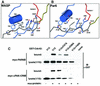
Fig. 5. (A) The interaction of the semi-CRIB motif of Par6 with its PDZ domain. As in Figure 3, dotted black lines represent salt bridges and hydrogen bonds. Non-sequence-specific hydrogen bonds (e.g., the main chain hydrogen bonds of the β-sheet) are not shown for clarity. (B) The interactions of Par6 with the Switch I and II regions of Cdc42. (C) Effect of Par6 and PAK GBD domains on the intrinsic GTPase activity of Cdc42. Cdc42 loaded with [γ-32P]GTP was incubated for 10 min plus saturating concentrations (20× _K_d) of the GST–GBD fusion proteins or GST alone. The Cdc42 was then bound to filters, washed to remove free 32P-phosphate and counted.
Fig. 6. Overlaid 1H/15N HSQC spectra of the Par6 PDZ domain (residues 152–253, red) and CRIB–PDZ elements (residues 126–253, black).
Fig. 7. Par6 B conformational biosensor. (A) Schematic of the biosensor structure, in the bound and unbound states, and emission spectra of the biosensor, CFP alone and YFP alone, excited at 433 nm. (B) Emission spectra of the CFP–Par6–YFP biosensor and of separated CFP and YFP. (C) Emission spectra of the biosensor and of CFP–YFP, each incubated with either Cdc42·GMPPNP or Cdc42·GDP (1 µM). (D) Titration curve of the Par6 biosensor incubated with varying concentrations of Q61L Cdc42 or the indicated mutants (in a Q61L background), all loaded with GMPPNP. The binding curve was fitted to the data assuming a single independent binding site, using Kaleidagraph software.
Similar articles
- Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition.
Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Peterson FC, et al. Mol Cell. 2004 Mar 12;13(5):665-76. doi: 10.1016/s1097-2765(04)00086-3. Mol Cell. 2004. PMID: 15023337 - Assembly of epithelial tight junctions is negatively regulated by Par6.
Gao L, Joberty G, Macara IG. Gao L, et al. Curr Biol. 2002 Feb 5;12(3):221-5. doi: 10.1016/s0960-9822(01)00663-7. Curr Biol. 2002. PMID: 11839275 - The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42.
Joberty G, Petersen C, Gao L, Macara IG. Joberty G, et al. Nat Cell Biol. 2000 Aug;2(8):531-9. doi: 10.1038/35019573. Nat Cell Biol. 2000. PMID: 10934474 - Cell polarity: the ups and downs of the Par6/aPKC complex.
Henrique D, Schweisguth F. Henrique D, et al. Curr Opin Genet Dev. 2003 Aug;13(4):341-50. doi: 10.1016/s0959-437x(03)00077-7. Curr Opin Genet Dev. 2003. PMID: 12888006 Review. - Flipping the switch: the structural basis for signaling through the CRIB motif.
Hoffman GR, Cerione RA. Hoffman GR, et al. Cell. 2000 Aug 18;102(4):403-6. doi: 10.1016/s0092-8674(00)00045-3. Cell. 2000. PMID: 10966102 Review. No abstract available.
Cited by
- The WD40 protein Morg1 facilitates Par6-aPKC binding to Crb3 for apical identity in epithelial cells.
Hayase J, Kamakura S, Iwakiri Y, Yamaguchi Y, Izaki T, Ito T, Sumimoto H. Hayase J, et al. J Cell Biol. 2013 Mar 4;200(5):635-50. doi: 10.1083/jcb.201208150. Epub 2013 Feb 25. J Cell Biol. 2013. PMID: 23439680 Free PMC article. - Always look on the bright site of Rho: structural implications for a conserved intermolecular interface.
Dvorsky R, Ahmadian MR. Dvorsky R, et al. EMBO Rep. 2004 Dec;5(12):1130-6. doi: 10.1038/sj.embor.7400293. EMBO Rep. 2004. PMID: 15577926 Free PMC article. Review. - The interaction of PTP-BL PDZ domains with RIL: an enigmatic role for the RIL LIM domain.
van den Berk LC, van Ham MA, te Lindert MM, Walma T, Aelen J, Vuister GW, Hendriks WJ. van den Berk LC, et al. Mol Biol Rep. 2004 Dec;31(4):203-15. doi: 10.1007/s11033-005-1407-8. Mol Biol Rep. 2004. PMID: 15663004 - Characterization of PDZ domain-peptide interaction interface based on energetic patterns.
Li N, Hou T, Ding B, Wang W. Li N, et al. Proteins. 2011 Nov;79(11):3208-20. doi: 10.1002/prot.23157. Epub 2011 Sep 17. Proteins. 2011. PMID: 21928318 Free PMC article. - Surface sites for engineering allosteric control in proteins.
Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R. Lee J, et al. Science. 2008 Oct 17;322(5900):438-42. doi: 10.1126/science.1159052. Science. 2008. PMID: 18927392 Free PMC article.
References
- Abdul-Manan N., Aghazadeh,B., Liu,G.A., Majumdar,A., Ouerfelli,O., Siminovitch,K.A. and Rosen,M.K. (1999) Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature, 399, 379–383. - PubMed
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
- Burbelo P.D., Drechsel,D. and Hall,A. (1995) A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J. Biol. Chem., 270, 29071–29074. - PubMed
- CCP4 (1994) The CCP4 suite of programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM070902/GM/NIGMS NIH HHS/United States
- CA40042/CA/NCI NIH HHS/United States
- GM56322/GM/NIGMS NIH HHS/United States
- P01 CA040042/CA/NCI NIH HHS/United States
- R01 GM056322/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous