The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection - PubMed (original) (raw)
The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection
Hiroki Isomura et al. J Virol. 2003 Mar.
Abstract
To determine the effect of the human cytomegalovirus (CMV) major immediate-early (MIE) enhancer or promoter on the efficiency of viral replication in permissive human cells, we constructed recombinant viruses with their human MIE promoter, enhancer, and promoter plus enhancer replaced with the murine CMV components. After a low multiplicity of infection (MOI) (0.01 PFU/cell), recombinant human CMV with the murine CMV promoter replicated like the wild type but recombinant virus with the murine enhancer replicated less efficiently. Immediate-early (IE) viral protein pIE72 (UL123), early viral protein (UL44), and viral DNA synthesis were significantly decreased. The effect of the human CMV enhancer substitution with the murine CMV enhancer was also demonstrated in different cell types by using recombinant virus with the UL127 promoter, driving the expression of green fluorescent protein (GFP). After an MOI of 1, GFP expression was high with the human CMV enhancer and significantly lower with the murine CMV enhancer. Even though at a high MOI (10 PFU/cell), the murine CMV enhancer was as efficient as the human CMV enhancer for the transcription of IE genes in human foreskin fibroblast cells, at lower MOIs, the murine CMV enhancer was less efficient. Proximal and distal chimeras of the human and murine enhancers also replicated less efficiently at a low MOI and expressed lower levels of GFP from the UL127 promoter. These experiments demonstrate that the entire human CMV enhancer has evolved for the efficient expression of the viral IE and early genes in human cells. Possible functions of the human CMV enhancer and promoter at a low MOI are discussed.
Figures
FIG. 1.
Structural analysis of recombinant viruses. (A) Schematic diagram of parental and recombinant viruses. All recombinant viruses containing GFP or CAT were derived from RVdlMSVgpt (parental virus). The murine CMV promoter (MCMVP), enhancer (MCMVE), and enhancer plus promoter (MCMVE/P) are designated with a bold arrow. The HCMV promoter (HCMVP), enhancer (HCMVE), and enhancer plus promoter are designated with a regular arrow. The viral DNA fragment sizes of all recombinant viruses digested with the restriction endonucleases _Blp_I and _Spe_I are indicated. 32P-Labeled probes for Southern blot hybridization to confirm the recombinations are shown at the bottom as probes 1, 2, and 3. Shuttle vector construction and viral DNA transfection of HFF cells are described in Materials and Methods. (B) Southern blot analysis of the parental virus and the recombinant viruses. Viral DNAs were digested with restriction endonucleases _Blp_I and _Spe_I, fractionated by electrophoresis in 1.0% agarose, and subjected to hybridization with 32P-labeled probe 1, 2, or 3. Standard molecular size markers are indicated to the left in base pairs. Lanes: 1, RVdlMSVgpt (parental virus); 2, RVdlMmP-GFP (mP-CAT); 3, RVdlMmE-GFP (mE-CAT); 4, RVdlMmE/P-GFP (mE/P-CAT); 5, RVdlMmP-CAT (mP-GFP); 6, RVdlMmE-CAT (mE-GFP); 7, RVdlMmE/P-CAT (mE/P-GFP).
FIG. 1.
Structural analysis of recombinant viruses. (A) Schematic diagram of parental and recombinant viruses. All recombinant viruses containing GFP or CAT were derived from RVdlMSVgpt (parental virus). The murine CMV promoter (MCMVP), enhancer (MCMVE), and enhancer plus promoter (MCMVE/P) are designated with a bold arrow. The HCMV promoter (HCMVP), enhancer (HCMVE), and enhancer plus promoter are designated with a regular arrow. The viral DNA fragment sizes of all recombinant viruses digested with the restriction endonucleases _Blp_I and _Spe_I are indicated. 32P-Labeled probes for Southern blot hybridization to confirm the recombinations are shown at the bottom as probes 1, 2, and 3. Shuttle vector construction and viral DNA transfection of HFF cells are described in Materials and Methods. (B) Southern blot analysis of the parental virus and the recombinant viruses. Viral DNAs were digested with restriction endonucleases _Blp_I and _Spe_I, fractionated by electrophoresis in 1.0% agarose, and subjected to hybridization with 32P-labeled probe 1, 2, or 3. Standard molecular size markers are indicated to the left in base pairs. Lanes: 1, RVdlMSVgpt (parental virus); 2, RVdlMmP-GFP (mP-CAT); 3, RVdlMmE-GFP (mE-CAT); 4, RVdlMmE/P-GFP (mE/P-CAT); 5, RVdlMmP-CAT (mP-GFP); 6, RVdlMmE-CAT (mE-GFP); 7, RVdlMmE/P-CAT (mE/P-GFP).
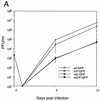
FIG. 2.
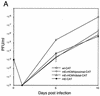
Growth kinetics of wt and recombinant viruses. (A) Multistep growth curve of wt and recombinant viruses at an MOI of 0.01. HFF cells were used for growth and the plaque assay of recombinant viruses wt-GFP, mP-GFP, mE-GFP, and mE/P-GFP as described in Materials and Methods. Four independent assays were used to determine the mean and the standard error. (B) Plaque size of wt and recombinant viruses. All plaques were generated from inoculums 5 d p.i. (a) wt-GFP; (b) mP-GFP; (c) mE-GFP; (d) mE/P-GFP. Magnification, ×0.5.
FIG. 2.
Growth kinetics of wt and recombinant viruses. (A) Multistep growth curve of wt and recombinant viruses at an MOI of 0.01. HFF cells were used for growth and the plaque assay of recombinant viruses wt-GFP, mP-GFP, mE-GFP, and mE/P-GFP as described in Materials and Methods. Four independent assays were used to determine the mean and the standard error. (B) Plaque size of wt and recombinant viruses. All plaques were generated from inoculums 5 d p.i. (a) wt-GFP; (b) mP-GFP; (c) mE-GFP; (d) mE/P-GFP. Magnification, ×0.5.
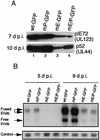
FIG. 3.
Early viral protein and DNA synthesis with wt and recombinant viruses. (A) Western blot of IE pIE72 (UL123) and early p52 (UL44) proteins after infection with wt or recombinant viruses at an MOI of 0.01. pIE72 (UL123) and p52 (UL44) were detected 7 and 10 d p.i. with monoclonal antibodies 810 and M0854, respectively, as described in Materials and Methods. (B) Analysis of viral DNA synthesis after infection of HFF cells with wt and recombinant viruses at an MOI of 0.01. DNAs from infected HFF cells were isolated 5 and 9 d p.i., digested with the restriction endonuclease _Hin_dIII, and subjected to Southern blot hybridization with either a 32P-labeled T probe or a lambda probe as described in Materials and Methods. Lambda DNA served as an internal control. Arrows designate the viral DNA fused ends (17.2 and 13.0 kb), free ends (9.7 kb), and the internal lambda DNA control.
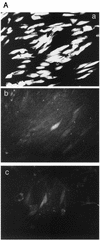
FIG. 4.
GFP expression from the UL127 promoter after infection of different cell types with wt and recombinant viruses. (A) GFP expression of HFF cells at 3 d p.i. at an MOI of 1 detected by fluorescence microscopy. (a) wt-GFP; (b) mE-GFP; (c) mE/P-GFP. Magnification, ×20. (B) GFP expression of THP-1 cells at 3 d p.i. at an MOI of 1. Differentiation of THP-1 cells was induced with 20 nM PMA plus 50 μM hydrocortisone. Mean fluorescence intensities were determined with a FACScan flow cytometer. Three independent experiments were used to determine the mean and the standard error.
FIG. 4.
GFP expression from the UL127 promoter after infection of different cell types with wt and recombinant viruses. (A) GFP expression of HFF cells at 3 d p.i. at an MOI of 1 detected by fluorescence microscopy. (a) wt-GFP; (b) mE-GFP; (c) mE/P-GFP. Magnification, ×20. (B) GFP expression of THP-1 cells at 3 d p.i. at an MOI of 1. Differentiation of THP-1 cells was induced with 20 nM PMA plus 50 μM hydrocortisone. Mean fluorescence intensities were determined with a FACScan flow cytometer. Three independent experiments were used to determine the mean and the standard error.
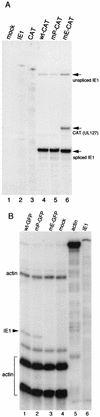
FIG. 5.
Steady-state mRNA levels transcribed from the MIE promoter or the UL127-CAT promoter. Cytoplasmic RNA was analyzed by an RNase protection assay at 6 (A) or 24 (B) h p.i. as described in Materials and Methods. (A) IE1 and UL127-CAT mRNA after a high MOI (approximately 5 PFU/cell) with cycloheximide. Probes used in lanes 2 and 3 lacked RNase T1. (B) IE1 mRNA after a low MOI (0.01 PFU/cell). Actin mRNA was used as a loading control. Probes used in lanes 5 and 6 lacked RNase T1.
FIG. 6.
Effect of different MOIs on IE transcription of wt-GFP and mE-GFP. Total cell RNAs and DNAs were isolated in parallel at 6 and 4 h p.i., respectively, and analyzed by multiplex real-time PCR as described in Materials and Methods. gB DNA or IE RNA was normalized to a internal ribosomal DNA or RNA control (respectively). (A) gB DNAs normalized (DNAn) relative to the wt with an MOI of 10, 1, or 0.1. (B) IE RNAs normalized (RNAn) relative to the wt with an MOI of 10, 1, 0.1 or 0.01.
FIG. 7.
Structural analysis of recombinant viruses with human CMV and murine CMV enhancer chimeras. (A) Schematic diagram of recombinant viruses. All recombinant viruses containing GFP or CAT were derived from RVdlMSVgpt (parental virus). Murine CMV components are designated with a bold arrow, and human CMV components are designated with a regular arrow (see the legend to Fig. 1A). The viral DNA fragment size of all recombinant viruses digested with restriction endonucleases _Blp_I and _Spe_I or _Pac_I and _Spe_I are indicated. 32P-Labeled probes for Southern blot hybridization to confirm the recombinations are shown at the bottom as probes 1 and 2. Shuttle vector construction and viral DNA transfection of HFF cells are described in Materials and Methods. (B) Southern blot analysis of the wt and recombinant viruses. Viral DNAs were digested with restriction endonucleases, fractionated by electrophoresis in 1.0% agarose, and subjected to hybridization with 32P-labeled probe 1 or 2. Standard molecular size markers are indicated in base pairs to the left of the two groups of gels.
FIG. 8.
Growth kinetics of the recombinant wt or human CMV and murine CMV enhancer chimeras. (A) Multistep growth curve of wt-CAT, mE+HCMVproximal-CAT, mE+HCMVdistal-CAT, or mE-CAT recombinant viruses at an MOI of 0.01. DNA was isolated in parallel and shown to be of equal input by multiplex real-time PCR as described in the legend to Fig. 6. HFF cells were used for growth and the plaque assay of the recombinant viruses as described in Materials and Methods. Three independent assays were used to determine the mean and the standard error. (B) Plaque sizes of parental and recombinant viruses. All plaques were generated from inoculums at 11 d p.i. (a) wt-CAT; (b) mE+HCMVproximal-CAT; (c) mE+HCMVdistal-CAT; (d) mE-CAT.
FIG. 8.
Growth kinetics of the recombinant wt or human CMV and murine CMV enhancer chimeras. (A) Multistep growth curve of wt-CAT, mE+HCMVproximal-CAT, mE+HCMVdistal-CAT, or mE-CAT recombinant viruses at an MOI of 0.01. DNA was isolated in parallel and shown to be of equal input by multiplex real-time PCR as described in the legend to Fig. 6. HFF cells were used for growth and the plaque assay of the recombinant viruses as described in Materials and Methods. Three independent assays were used to determine the mean and the standard error. (B) Plaque sizes of parental and recombinant viruses. All plaques were generated from inoculums at 11 d p.i. (a) wt-CAT; (b) mE+HCMVproximal-CAT; (c) mE+HCMVdistal-CAT; (d) mE-CAT.
FIG. 9.
Effect of proximal or distal human CMV and murine CMV enhancer chimeras on GFP expression from the UL127 promoter. HFF cells were infected at an MOI of 1, and GFP expression was detected by fluorescence microscopy at 3 d p.i. (a) wt-GFP; (b) mE+HCMVproximal-GFP; (c) mE+HCMVdistal-GFP; (d) mE-GFP. Magnification, ×20.
Similar articles
- The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression.
Meier JL, Pruessner JA. Meier JL, et al. J Virol. 2000 Feb;74(4):1602-13. doi: 10.1128/jvi.74.4.1602-1613.2000. J Virol. 2000. PMID: 10644329 Free PMC article. - A strong negative transcriptional regulatory region between the human cytomegalovirus UL127 gene and the major immediate-early enhancer.
Lundquist CA, Meier JL, Stinski MF. Lundquist CA, et al. J Virol. 1999 Nov;73(11):9039-52. doi: 10.1128/JVI.73.11.9039-9052.1999. J Virol. 1999. PMID: 10516010 Free PMC article. - Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency.
Stinski MF, Isomura H. Stinski MF, et al. Med Microbiol Immunol. 2008 Jun;197(2):223-31. doi: 10.1007/s00430-007-0069-7. Epub 2007 Dec 19. Med Microbiol Immunol. 2008. PMID: 18097687 Review. - Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication.
Isomura H, Tsurumi T, Stinski MF. Isomura H, et al. J Virol. 2004 Dec;78(23):12788-99. doi: 10.1128/JVI.78.23.12788-12799.2004. J Virol. 2004. PMID: 15542631 Free PMC article. - Coordination of late gene transcription of human cytomegalovirus with viral DNA synthesis: recombinant viruses as potential therapeutic vaccine candidates.
Isomura H, Stinski MF. Isomura H, et al. Expert Opin Ther Targets. 2013 Feb;17(2):157-66. doi: 10.1517/14728222.2013.740460. Epub 2012 Dec 12. Expert Opin Ther Targets. 2013. PMID: 23231449 Review.
Cited by
- Human cytomegalovirus inhibition by cardiac glycosides: evidence for involvement of the HERG gene.
Kapoor A, Cai H, Forman M, He R, Shamay M, Arav-Boger R. Kapoor A, et al. Antimicrob Agents Chemother. 2012 Sep;56(9):4891-9. doi: 10.1128/AAC.00898-12. Epub 2012 Jul 9. Antimicrob Agents Chemother. 2012. PMID: 22777050 Free PMC article. - In vivo competence of murine cytomegalovirus under the control of the human cytomegalovirus major immediate-early enhancer in the establishment of latency and reactivation.
Gustems M, Busche A, Messerle M, Ghazal P, Angulo A. Gustems M, et al. J Virol. 2008 Oct;82(20):10302-7. doi: 10.1128/JVI.01255-08. Epub 2008 Aug 6. J Virol. 2008. PMID: 18684819 Free PMC article. - Prolonged activation of NF-kappaB by human cytomegalovirus promotes efficient viral replication and late gene expression.
DeMeritt IB, Podduturi JP, Tilley AM, Nogalski MT, Yurochko AD. DeMeritt IB, et al. Virology. 2006 Mar 1;346(1):15-31. doi: 10.1016/j.virol.2005.09.065. Epub 2005 Nov 21. Virology. 2006. PMID: 16303162 Free PMC article. - RNA-targeted proteomics identifies YBX1 as critical for efficient HCMV mRNA translation.
Dickmander RJ, Lenarcic EM, Sears JD, Hale AE, Moorman NJ. Dickmander RJ, et al. Proc Natl Acad Sci U S A. 2025 Mar 11;122(10):e2421155122. doi: 10.1073/pnas.2421155122. Epub 2025 Mar 4. Proc Natl Acad Sci U S A. 2025. PMID: 40035757 Free PMC article. - Role of the human cytomegalovirus major immediate-early promoter's 19-base-pair-repeat cyclic AMP-response element in acutely infected cells.
Keller MJ, Wheeler DG, Cooper E, Meier JL. Keller MJ, et al. J Virol. 2003 Jun;77(12):6666-75. doi: 10.1128/jvi.77.12.6666-6675.2003. J Virol. 2003. PMID: 12767986 Free PMC article.
References
- Anders, D. G., and L. A. McCue. 1996. The human cytomegalovirus genes and proteins required for DNA synthesis. Intervirology 39:378-388. - PubMed
- Angulo, A., C. Suto, R. A. Heyman, and P. Ghazal. 1995. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol. Endocrinol. 10:781-793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources