Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex - PubMed (original) (raw)
Comparative Study
. 2003 Mar;23(6):1863-73.
doi: 10.1128/MCB.23.6.1863-1873.2003.
Yuki Yamaguchi, Keiichi Yano, Seiji Sugimoto, Sittinan Chanarat, Tadashi Wada, Dong-ki Kim, Jun Hasegawa, Masashi Omori, Naoto Inukai, Masaki Endoh, Tomoko Yamada, Hiroshi Handa
Affiliations
- PMID: 12612062
- PMCID: PMC149481
- DOI: 10.1128/MCB.23.6.1863-1873.2003
Comparative Study
Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex
Takashi Narita et al. Mol Cell Biol. 2003 Mar.
Abstract
The multisubunit transcription elongation factor NELF (for negative elongation factor) acts together with DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) sensitivity-inducing factor (DSIF)/human Spt4-Spt5 to cause transcriptional pausing of RNA polymerase II (RNAPII). NELF activity is associated with five polypeptides, A to E. NELF-A has sequence similarity to hepatitis delta antigen (HDAg), the viral protein that binds to and activates RNAPII, whereas NELF-E is an RNA-binding protein whose RNA-binding activity is critical for NELF function. To understand the interactions of DSIF, NELF, and RNAPII at a molecular level, we identified the B, C, and D proteins of human NELF. NELF-B is identical to COBRA1, recently reported to associate with the product of breast cancer susceptibility gene BRCA1. NELF-C and NELF-D are highly related or identical to the protein called TH1, of unknown function. NELF-B and NELF-C or NELF-D are integral subunits that bring NELF-A and NELF-E together, and coexpression of these four proteins in insect cells resulted in the reconstitution of functionally active NELF. Detailed analyses using mutated recombinant complexes indicated that the small region of NELF-A with similarity to HDAg is critical for RNAPII binding and for transcriptional pausing. This study defines several important protein-protein interactions and opens the way for understanding the mechanism of DSIF- and NELF-induced transcriptional pausing.
Figures
FIG. 1.
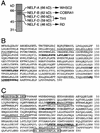
Cloning of NELF-B, NELF-C, and NELF-D. (A) NELF was purified from human HeLa cell nuclear extracts as described previously (37). NELF from the final purification step (mono S) was visualized by silver staining. (B) Amino acid sequence of human NELF-B. The three obtained peptide sequences are shown in boldface. Dotted lines indicate two regions predicted (probability, >0.5) by a 21-residue scan with the program COILS to form the coiled-coil structure. (C) Amino acid sequence of human NELF-C and NELF-D. The three peptide sequences obtained from NELF-C are shown in boldface. The four peptide sequences obtained from NELF-D are underlined. Boxed are the sequences used for the generation of antipeptide antibodies N12 and C14. Amino acids in brackets are probably unique to NELF-C.
FIG. 2.
Distinction between NELF-C and NELF-D. (A) A cDNA encoding the 590-aa form of TH1 was cloned under the T3 promoter and used as a template for in vitro transcription and translation. Translation was performed with rabbit reticulocyte lysates in the presence of [35S]cysteine, and reaction products were analyzed by SDS-PAGE and autoradiography. The 5′-terminal sequence of the cDNA and putative translation initiation sites are shown. (B) Affinity-purified NELF (affi. NELF) and the 581- and 590-aa forms of recombinant TH1 proteins with histidine tags (His) were examined by immunoblotting (IB) with the antibodies N12 and C14. (C) NELF affinity purified from a HeLa cell line constitutively expressing Flag-E was immunoprecipitated (IP) with the antibodies N12 and c-Myc (control). Bound materials and 10% of the input were analyzed by SDS-PAGE and silver staining. The asterisk denotes the position of immunoglobulin heavy chains. Though bands of NELF-A and NELF-E are not clearly visible in lane 1 on this gel, their presence has been confirmed by immunoblot analysis (data not shown).
FIG. 3.
NELF subunits are ubiquitously expressed in various human tissues. Multiple-tissue Northern blot (Clontech) containing 2 μg of poly(A) -containing mRNA per lane was used to determine expression levels of NELF subunit mRNAs in a variety of human tissues. Appropriate restriction fragments of NELF subunit cDNAs were gel purified and 32P labeled by using Klenow DNA polymerase and random hexamers. Human β-actin probe (Clontech) was also used to confirm equivalent loadings of intact RNA. The positions of markers in kilobases are indicated to the left.
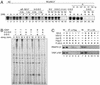
FIG. 4.
Reconstitution of NELF complex with recombinant proteins. (A) pFastBac (pFB)-based DNAs constructed for expression of recombinant NELF subunits. Each subunit was fused to one of four epitope tags, as shown. (B) Coomassie blue staining of purified recombinant NELF (rNELF). Sf9 cells that were multiply infected by recombinant baculoviruses for HA-A, Myc-B, VSV-D, and Flag-E were lysed and subjected to anti-Flag affinity chromatography as described in Materials and Methods. Purified proteins were resolved by SDS-7.5% PAGE and stained by Coomassie blue. The identities of these bands were determined by using anti-epitope tag antibodies and are indicated to the right. The positions of molecular mass markers in kilodaltons (kD) are indicated to the left. (C) Identification of multiple interactions among NELF subunits. Sf9 cells were infected by the recombinant baculoviruses in various combinations. These cells were lysed, immunoprecipitated (IP) with anti-Flag (α-Flag) (lanes 1 to 5), anti-HA (α-HA) (lanes 6 to 9), and anti-VSV (α-VSV) (lanes 10 and 11) antibodies, and analyzed by immunoblotting (IB) with the indicated antibodies. (D) Summary of the results. (Top) O's and X's indicate the presence and absence of interactions, respectively. (Bottom) Schematic structure of NELF complex.
FIG. 5.
Functional analyses of recombinant NELF and its subassemblies. (A) Promoter- and factor-dependent transcription with HeLa nuclear extracts (NE). Supercoiled plasmid pTF3-6C2AT (25 ng) and either untreated (lanes 1 and 2) or NELF-immunodepleted (NEΔNELF) (lanes 3 to 26) nuclear extracts (2 μl) were incubated in the presence or absence of DRB (50 μM). NELF affinity purified (affi. NELF) from a HeLa cell line constitutively expressing Flag-E, recombinant NELF complexes containing various numbers of subunits, and bacterially expressed NELF-A were added to the reaction mixtures where indicated. The notations 0.5×, 1.5×, 2×, 4×, and 6× indicate the amounts of exogenously added NELF relative to that of endogenous NELF contained in 2-μl untreated nuclear extracts (i.e., 10 ng of NELF-B protein). Full-length (380-nucleotide [nt]) G-free transcripts are shown. (B) Promoter- and factor-independent transcription with RNAPII. dC-tailed template (25 fmol) and purified RNAPII (25 fmol) were incubated. Recombinant DSIF (rDSIF) (p160, 30 ng; p14, 3 ng) and recombinant NELF complexes containing 12 ng of NELF-B were included where indicated. Nucleoside triphosphates were added, and elongation (elong.) was allowed to proceed for 4 and 16 min. Note that short transcripts of <20 nt flowed through the 6% polyacrylamide gel containing 8 M urea. (C) Binding of recombinant NELF to DSIF and RNAPII. Sf9 cells infected with various combinations of the recombinant baculoviruses were lysed and incubated with P.3/D.3, a HeLa nuclear extract-derived fraction rich in DSIF and RNAPII but devoid of NELF. The mixtures were immunoprecipitated (IP) with anti-Flag (α-Flag) (lanes 2 to 6) and anti-HA (α-HA) (lanes 8 to 12) antibodies. The presence of DSIF and RNAPII was determined by immunoblotting with anti-DSIF p160 and anti-RNAPII large subunit (LS) antibodies. Input (IN) lanes represent 2% of P.3/D.3 used for IP.
FIG. 6.
NELF-A interacts with NELF-C or NELF-D through a short segment in the middle of the HDAg homology region. Various fragments of NELF-A were expressed as GST fusion proteins (GST-A) in E. coli. These proteins (∼1 μg) were coupled to glutathione Sepharose and incubated with NELF-D protein that was synthesized and 35S labeled in vitro. Bound materials as well as 50% of the input (IN) were analyzed by SDS-PAGE and autoradiography. +++, strong (>25%) binding; +, weak (5 to 10%) binding; +/−, very weak (2 to 5%) binding; −, no (<2%) binding. HDAg-hom., the region of NELF-A with similarity to HDAg; PST, the region rich in proline, serine, and threonine residues.
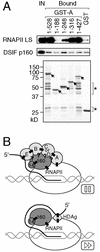
FIG. 7.
A part of the NELF-A region with similarity to HDAg (HDAg-hom.) is important for RNAPII binding and for transcriptional repression. (A) C-terminally truncated mutants of NELF-A used in this study. The region responsible for NELF-C or NELF-D binding is indicated at the top. A summary of the results presented in panels C and D is shown to the right. +++, comparable to wild-type activity; ++, slightly weaker than wild-type activity; −, significantly weaker than wild-type activity. PST, the region rich in proline, serine, and threonine residues. (B) Recombinant NELF (rNELF) complexes containing wild-type and mutant NELF-As were purified by anti-Flag affinity chromatography and analyzed by SDS-12.5% PAGE and Coomassie blue staining. Arrowheads indicate the positions of wild-type and mutant NELF-As. (C) Transcriptional repression by the rNELF mutants. pTF3-6C2AT (25 ng) and NELF-depleted nuclear extracts (NEΔNELF; 2 μl) were incubated in the presence of DRB (50 μM). NELF affinity purified (affi. NELF) from a HeLa cell line constitutively expressing Flag-E (lane 2) and rNELF complexes containing wild-type and mutant NELF-As (lanes 3 to 12) were included in the reaction mixtures where indicated. The notations 1× and 3× indicate the amounts of exogenously added NELF relative to that of endogenous NELF contained in 2-μl untreated nuclear extracts (i.e., 10 ng of NELF-B protein). The counts of 380-nt G-free transcripts were quantified by using the image analyzer STORM (Molecular Dynamics). Transcription levels relative to that of lane 1 (the reaction without NELF) are shown at the bottom. (D) Binding of the rNELF mutants to DSIF and RNAPII. Binding assays were carried out as described in the legend for Fig. 5C. IN, input; IP, immunoprecipitate; α-Flag, anti-Flag antibody; LS, large subunit.
FIG. 8.
Evidence that NELF-A binds directly to RNAPII. (A) The same NELF-A mutant proteins used in the analyses described in Fig. 7 were expressed as GST fusion proteins (GST-As). These proteins (∼1 μg) were coupled to glutathione Sepharose and incubated with P.3/D.3 as described in Materials and Methods. Bound materials as well as 2% of the input (IN) were analyzed by SDS-PAGE and either immunoblotting (top) or Coomassie blue staining (bottom). Arrows indicate the expected sizes of GST-As. Most of the smaller polypeptides visible on the gel (indicated by asterisks) originated from bacterial lysates and probably represent the C-terminally truncated forms of GST-As. LS, large subunit. (B) Possible model for DSIF- and NELF-induced transcriptional pausing (top). As a comparison, a model for the HDAg-activated elongation is shown at the bottom. See the text for details. Subunits of NELF and DSIF are shown in light gray and dark gray, respectively. Filled boxes indicate experimentally proven interactions, while question marks indicate hypothetical interactions. Interactions of DSIF p160/Spt5 with DSIF p14/Spt4 and RNAPII were demonstrated previously (9, 36). RNA bindings by NELF-E and HDAg were shown previously (16, 40). RNAPII binding by HDAg was also shown previously (38).
Similar articles
- Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA.
Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Yamaguchi Y, et al. Mol Cell Biol. 2002 May;22(9):2918-27. doi: 10.1128/MCB.22.9.2918-2927.2002. Mol Cell Biol. 2002. PMID: 11940650 Free PMC article. - Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex.
Missra A, Gilmour DS. Missra A, et al. Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11301-6. doi: 10.1073/pnas.1000681107. Epub 2010 Jun 4. Proc Natl Acad Sci U S A. 2010. PMID: 20534440 Free PMC article. - Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond.
Yamaguchi Y, Shibata H, Handa H. Yamaguchi Y, et al. Biochim Biophys Acta. 2013 Jan;1829(1):98-104. doi: 10.1016/j.bbagrm.2012.11.007. Epub 2012 Nov 29. Biochim Biophys Acta. 2013. PMID: 23202475 Review. - HIV-1 Tat: coping with negative elongation factors.
Garber ME, Jones KA. Garber ME, et al. Curr Opin Immunol. 1999 Aug;11(4):460-5. doi: 10.1016/S0952-7915(99)80077-6. Curr Opin Immunol. 1999. PMID: 10448148 Review.
Cited by
- Translational Initiation at a Non-AUG Start Codon for Human and Mouse Negative Elongation Factor-B.
Pan H, Zhao X, Zhang X, Abouelsoud M, Sun J, April C, Amleh A, Fan JB, Hu Y, Li R. Pan H, et al. PLoS One. 2015 May 26;10(5):e0127422. doi: 10.1371/journal.pone.0127422. eCollection 2015. PLoS One. 2015. PMID: 26010750 Free PMC article. - Role of RNA Polymerase II Promoter-Proximal Pausing in Viral Transcription.
Whelan M, Pelchat M. Whelan M, et al. Viruses. 2022 Sep 13;14(9):2029. doi: 10.3390/v14092029. Viruses. 2022. PMID: 36146833 Free PMC article. Review. - DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation.
Zhu W, Wada T, Okabe S, Taneda T, Yamaguchi Y, Handa H. Zhu W, et al. Nucleic Acids Res. 2007;35(12):4064-75. doi: 10.1093/nar/gkm430. Epub 2007 Jun 12. Nucleic Acids Res. 2007. PMID: 17567605 Free PMC article. - The super elongation complex (SEC) mediates phase transition of SPT5 during transcriptional pause release.
Guo C, Zhang Y, Shuai S, Sigbessia A, Hao S, Xie P, Jiang X, Luo Z, Lin C. Guo C, et al. EMBO Rep. 2023 Mar 6;24(3):e55699. doi: 10.15252/embr.202255699. Epub 2023 Jan 11. EMBO Rep. 2023. PMID: 36629390 Free PMC article. - Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator.
Malik S, Barrero MJ, Jones T. Malik S, et al. Proc Natl Acad Sci U S A. 2007 Apr 10;104(15):6182-7. doi: 10.1073/pnas.0608717104. Epub 2007 Apr 2. Proc Natl Acad Sci U S A. 2007. PMID: 17404243 Free PMC article.
References
- Bonthron, D. T., B. E. Hayward, V. Moran, and L. Strain. 2000. Characterization of TH1 and CTSZ, two non-imprinted genes downstream of GNAS1 in chromosome 20q13. Hum. Genet. 107:165-175. - PubMed
- Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. - PubMed
- Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous