EMF genes maintain vegetative development by repressing the flower program in Arabidopsis - PubMed (original) (raw)
Comparative Study
EMF genes maintain vegetative development by repressing the flower program in Arabidopsis
Yong-Hwan Moon et al. Plant Cell. 2003 Mar.
Erratum in
- Plant Cell. 2003 May;15(5):1257
Abstract
The EMBRYONIC FLOWER (EMF) genes EMF1 and EMF2 are required to maintain vegetative development and repress flower development. EMF1 encodes a putative transcriptional regulator, and EMF2 encodes a Polycomb group protein homolog. We examined expression profiles of emf mutants using GeneChip technology. The high degree of overlap in expression changes from the wild type among the emf1 and emf2 mutants was consistent with the functional similarity between the two genes. Expression profiles of emf seedlings before flower development were similar to that of Arabidopsis flowers, indicating the commitment of germinating emf seedlings to the reproductive fate. The germinating emf seedlings ectopically expressed flower organ genes, suggesting that vegetative development in wild-type plants results from EMF repression of the flower program, directly or indirectly. In addition, the seed development program is derepressed in the emf1 mutants. Gene expression analysis showed no clear regulation of CONSTANS (CO), FLOWERING LOCUS T (FT), LEAFY (LFY), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 by EMF1. Consistent with epistasis results that co, lfy, or ft cannot rescue rosette development in emf mutants, these data show that the mechanism of EMF-mediated repression of flower organ genes is independent of these flowering genes. Based on these findings, a new mechanism of EMF-mediated floral repression is proposed.
Figures
Figure 1.
Phenotypes of Wild-Type and emf Mutants. Seven-day-old wild-type (WT) Columbia ecotype (A), emf1-1 (B), emf1-2 (C), and emf2-1 (D), 15-day-old wild-type Columbia (E), emf1-1 (F), emf1-2 (G), and emf2-1 (H), and 21-day-old wild-type Columbia (I), emf1-1 (J), emf1-2 (K), and emf2-1 (L). A Columbia ecotype flower cluster is shown in (M). Bars = 1 mm in (A) to (D) and (F) to (H), 1.4 mm in (E), 1.5 mm in (I), 1.1 mm in (J) to (L), and 3.8 mm in (M).
Figure 2.
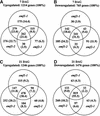
Genes Upregulated or Downregulated in 7- and 21-Day-Old emf1-1, emf1-2, and emf2-1 Mutants Relative to the Wild Type. From the GeneChip data, 3872 and 3820 genes with a hybridization signal of >50 in 7- and 21-day-old plants, respectively, were selected and analyzed. Numbers and percentages (in parentheses) of genes upregulated at 7 (A) and 21 (C) DAG and downregulated at 7 (B) and 21 (D) DAG in a mutant with greater than twofold difference are shown.
Figure 3.
Temporal Expression Patterns of the Flowering Genes and Seed Maturation Genes in the Wild Type and emf Mutants. Horizontal and vertical axes represent days after germination and intensities of the hybridization signal, respectively. See Tables 3 and 4 for the intensity of each gene's hybridization signal. WT, wild type.
Figure 4.
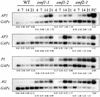
Expression of AP1, AP3, PI, and AG in emf Mutants. Autoradiograph of semiquantitative RT-PCR analysis of AP1, AP3, PI, AG, and GAPc RNA levels in 4-, 7-, 14-, and 21-day-old wild-type (WT) Columbia, emf1-1, emf1-2, and emf2-1 plants grown under SD conditions. GAPc was used as a RT-PCR control. RT-PCR products amplified were hybridized with a probe of each gene. The relative amount of each gene after standardization using the GAPc signal as a reference is presented below each gel. FC, flower clusters consisting of inflorescence meristems and floral buds.
Figure 5.
Expression of CO, FT, SOC1, and LFY in emf Mutants. Autoradiograph of semiquantitative RT-PCR analysis of CO, FT, SOC1, LFY, and GAPc RNA levels in 4-, 7-, 14-, and 21-day-old wild-type (WT) Columbia, emf1-1, emf1-2, and emf2-1 plants grown under SD conditions. GAPc was used as a RT-PCR control. RT-PCR products amplified were hybridized with a probe of each gene. The relative amount of each gene after standardization using the GAPc signal as a reference is presented below each gel. FC, flower clusters consisting of inflorescence meristems and floral buds.
Figure 6.
Histochemical Localization of GUS Activity in 7- and 14-Day-Old Wild-Type and emf1 Seedlings Harboring the LFY::GUS or AP1:: GUS Transgene. GUS activity is indicated by blue color. Bars = 0.5 mm. (A) LFY::GUS wild type (WT) at 7 DAG, showing GUS activity at the shoot tip (arrow). (B) LFY::GUS wild type at 14 DAG, showing GUS activity at the meristematic region of the shoot tip (arrow). (C) LFY::GUS emf1-1 at 7 DAG, showing GUS activity at the shoot tip (arrow). (D) LFY::GUS emf1-1 at 14 DAG, showing GUS activity at the shoot tip. (E) LFY::GUS emf1-2 at 7 DAG, showing GUS activity at the shoot tip (arrow). (F) LFY::GUS emf1-2 at 14 DAG, showing GUS activity in a patch of the carpelloid tissue (arrow). (G) AP1::GUS emf1-2 at 7 DAG, showing GUS activity in the shoot tip, cotyledons, and hypocotyl. (H) AP1::GUS emf1-2 at 14 DAG, showing GUS activity in the carpelloid structure and the papillae tissue developed at the base of the cotyledons (arrows).
Similar articles
- Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis.
Kim SY, Zhu T, Sung ZR. Kim SY, et al. Plant Physiol. 2010 Feb;152(2):516-28. doi: 10.1104/pp.109.143495. Epub 2009 Sep 25. Plant Physiol. 2010. PMID: 19783648 Free PMC article. - Temporal and spatial requirement of EMF1 activity for Arabidopsis vegetative and reproductive development.
Sánchez R, Kim MY, Calonje M, Moon YH, Sung ZR. Sánchez R, et al. Mol Plant. 2009 Jul;2(4):643-653. doi: 10.1093/mp/ssp004. Epub 2009 Mar 24. Mol Plant. 2009. PMID: 19825645 - EMF genes interact with late-flowering genes in regulating floral initiation genes during shoot development in Arabidopsis thaliana.
Chou ML, Haung MD, Yang CH. Chou ML, et al. Plant Cell Physiol. 2001 May;42(5):499-507. doi: 10.1093/pcp/pce062. Plant Cell Physiol. 2001. PMID: 11382816 - [Regulation network and biological roles of LEAFY in Arabidopsis thaliana in floral development].
Wang LL, Liang HM, Pang JL, Zhu MY. Wang LL, et al. Yi Chuan. 2004 Jan;26(1):137-42. Yi Chuan. 2004. PMID: 15626683 Review. Chinese. - Molecular mechanisms of floral organ specification by MADS domain proteins.
Yan W, Chen D, Kaufmann K. Yan W, et al. Curr Opin Plant Biol. 2016 Feb;29:154-62. doi: 10.1016/j.pbi.2015.12.004. Epub 2016 Jan 21. Curr Opin Plant Biol. 2016. PMID: 26802807 Review.
Cited by
- LEAFY COTYLEDONs: Connecting different stages of plant development.
Chen C, Du X. Chen C, et al. Front Plant Sci. 2022 Aug 31;13:916831. doi: 10.3389/fpls.2022.916831. eCollection 2022. Front Plant Sci. 2022. PMID: 36119568 Free PMC article. Review. - Knowing When to Silence: Roles of Polycomb-Group Proteins in SAM Maintenance, Root Development, and Developmental Phase Transition.
Yan B, Lv Y, Zhao C, Wang X. Yan B, et al. Int J Mol Sci. 2020 Aug 15;21(16):5871. doi: 10.3390/ijms21165871. Int J Mol Sci. 2020. PMID: 32824274 Free PMC article. Review. - Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis.
Wang D, Tyson MD, Jackson SS, Yadegari R. Wang D, et al. Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13244-9. doi: 10.1073/pnas.0605551103. Epub 2006 Aug 21. Proc Natl Acad Sci U S A. 2006. PMID: 16924116 Free PMC article. - H2AK121ub in Arabidopsis associates with a less accessible chromatin state at transcriptional regulation hotspots.
Yin X, Romero-Campero FJ, de Los Reyes P, Yan P, Yang J, Tian G, Yang X, Mo X, Zhao S, Calonje M, Zhou Y. Yin X, et al. Nat Commun. 2021 Jan 12;12(1):315. doi: 10.1038/s41467-020-20614-1. Nat Commun. 2021. PMID: 33436613 Free PMC article. - Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis.
Kim SY, Zhu T, Sung ZR. Kim SY, et al. Plant Physiol. 2010 Feb;152(2):516-28. doi: 10.1104/pp.109.143495. Epub 2009 Sep 25. Plant Physiol. 2010. PMID: 19783648 Free PMC article.
References
- Araki, T. (2001). Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 4, 63–68. - PubMed
- Bai, S., Chen, L., Yund, M.A., and Sung, Z.R. (2000). Mechanisms of plant embryo development. Curr. Top. Dev. Biol. 50, 61–88. - PubMed
- Bai, S., and Sung, Z.R. (1995). The role of EMF1 in regulating the vegetative and reproductive transition in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 82, 1095–1103.
- Birve, A., Sengupta, A.K., Beuchle, D., Larsson, J., Kennison, J.A., Rasmuson-Lestander, A., and Muller, J. (2001). Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128, 3371–3379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials