The genetic core of the universal ancestor - PubMed (original) (raw)
The genetic core of the universal ancestor
J Kirk Harris et al. Genome Res. 2003 Mar.
Abstract
Molecular analysis of conserved sequences in the ribosomal RNAs of modern organisms reveals a three-domain phylogeny that converges in a universal ancestor for all life. We used the Clusters of Orthologous Groups database and information from published genomes to search for other universally conserved genes that have the same phylogenetic pattern as ribosomal RNA, and therefore constitute the ancestral genetic core of cells. Our analyses identified a small set of genes that can be traced back to the universal ancestor and have coevolved since that time. As indicated by earlier studies, almost all of these genes are involved with the transfer of genetic information, and most of them directly interact with the ribosome. Other universal genes have either undergone lateral transfer in the past, or have diverged so much in sequence that their distant past could not be resolved. The nature of the conserved genes suggests innovations that may have been essential to the divergence of the three domains of life. The analysis also identified several genes of unknown function with phylogenies that track with the ribosomal RNA genes. The products of these genes are likely to play fundamental roles in cellular processes.
Figures
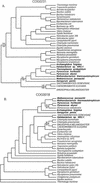
Figure 1.
Examples of three-domain and non-three-domain phylogenetic trees from analyses of the COG database protein alignments. The trees are the single shortest trees found by a maximum parsimony (MP) analysis of the amino acid alignments (neighbor-joining [NJ] analysis gave the same topology). Names of organisms belonging to the Bacteria are in italics; names of Archaea are in bold italics, and names of Eucarya are in all capital letters. (A) The phylogeny of COG0231 (efp in E. coli) recapitulates the basic three-domain topology given by ribosomal RNA, and the numbers indicate bootstrap support for the monophyly of the archaeal, bacterial, and eucaryal sequences in this COG. Results from MP bootstrap analysis are given above the branches, and results from NJ bootstrap analysis are given below. (B) The phylogeny of COG0018 (argS in E. coli) violates the three-domain paradigm, as none of the three domains are monophyletic. Indications of horizontal gene transfer events are presented in enlarged font along with corresponding bootstrap support for the nodes demarking lateral transfer.
Similar articles
- Protein content of minimal and ancestral ribosome.
Mushegian A. Mushegian A. RNA. 2005 Sep;11(9):1400-6. doi: 10.1261/rna.2180205. Epub 2005 Jul 25. RNA. 2005. PMID: 16043494 Free PMC article. - The origins of modern proteomes.
Kurland CG, Canbäck B, Berg OG. Kurland CG, et al. Biochimie. 2007 Dec;89(12):1454-63. doi: 10.1016/j.biochi.2007.09.004. Epub 2007 Sep 15. Biochimie. 2007. PMID: 17949885 - The structure and evolution of the ribosomal proteins encoded in the spc operon of the archaeon (Crenarchaeota) Sulfolobus acidocaldarius.
Yang D, Kusser I, Köpke AK, Koop BF, Matheson AT. Yang D, et al. Mol Phylogenet Evol. 1999 Jul;12(2):177-85. doi: 10.1006/mpev.1998.0607. Mol Phylogenet Evol. 1999. PMID: 10381320 - A genomic perspective on protein families.
Tatusov RL, Koonin EV, Lipman DJ. Tatusov RL, et al. Science. 1997 Oct 24;278(5338):631-7. doi: 10.1126/science.278.5338.631. Science. 1997. PMID: 9381173 Review. - Molecular evolution before the origin of species.
Davis BK. Davis BK. Prog Biophys Mol Biol. 2002 May-Jul;79(1-3):77-133. doi: 10.1016/s0079-6107(02)00012-3. Prog Biophys Mol Biol. 2002. PMID: 12225777 Review.
Cited by
- Phylogenomic Analyses and Comparative Studies on Genomes of the Bifidobacteriales: Identification of Molecular Signatures Specific for the Order Bifidobacteriales and Its Different Subclades.
Zhang G, Gao B, Adeolu M, Khadka B, Gupta RS. Zhang G, et al. Front Microbiol. 2016 Jun 27;7:978. doi: 10.3389/fmicb.2016.00978. eCollection 2016. Front Microbiol. 2016. PMID: 27446019 Free PMC article. - The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse?
Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. Gribaldo S, et al. Nat Rev Microbiol. 2010 Oct;8(10):743-52. doi: 10.1038/nrmicro2426. Nat Rev Microbiol. 2010. PMID: 20844558 Review. - Structural and functional divergence within the Dim1/KsgA family of rRNA methyltransferases.
Pulicherla N, Pogorzala LA, Xu Z, O Farrell HC, Musayev FN, Scarsdale JN, Sia EA, Culver GM, Rife JP. Pulicherla N, et al. J Mol Biol. 2009 Sep 4;391(5):884-93. doi: 10.1016/j.jmb.2009.06.015. Epub 2009 Jun 9. J Mol Biol. 2009. PMID: 19520088 Free PMC article. - The origin of modern terrestrial life.
Forterre P, Gribaldo S. Forterre P, et al. HFSP J. 2007 Sep;1(3):156-68. doi: 10.2976/1.2759103. Epub 2007 Jul 25. HFSP J. 2007. PMID: 19404443 Free PMC article. - Mycobacterial Membrane Proteins QcrB and AtpE: Roles in Energetics, Antibiotic Targets, and Associated Mechanisms of Resistance.
Bown L, Srivastava SK, Piercey BM, McIsaac CK, Tahlan K. Bown L, et al. J Membr Biol. 2018 Feb;251(1):105-117. doi: 10.1007/s00232-017-9997-3. Epub 2017 Nov 2. J Membr Biol. 2018. PMID: 29098330
References
- Aiyar A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132: 221-241. - PubMed
- Ban N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905-920. - PubMed
- Brown J.R., Douady, C.J., Italia, M.J., Marshall, W.E., and Stanhope, M.J. 2001b. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28: 281-285. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous