Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease - PubMed (original) (raw)
Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease
Mary M Brinig et al. Appl Environ Microbiol. 2003 Mar.
Abstract
Members of the uncultivated bacterial division TM7 have been detected in the human mouth, but little information is available regarding their prevalence and diversity at this site. Human subgingival plaque samples from healthy sites and sites exhibiting various stages of periodontal disease were analyzed for the presence of TM7 bacteria. TM7 ribosomal DNA (rDNA) was found in 96% of the samples, and it accounted for approximately 0.3%, on average, of all bacterial rDNA in the samples as determined by real-time quantitative PCR. Two new phylotypes of this division were identified, and members of the division were found to exhibit filamentous morphology by fluorescence in situ hybridization. The abundance of TM7 rDNA relative to total bacterial rDNA was higher in sites with mild periodontitis (0.54% +/- 0.1%) than in either healthy sites (0.21% +/- 0.05%, P < 0.01) or sites with severe periodontitis (0.29% +/- 0.06%, P < 0.05). One division subgroup, the I025 phylotype, was detected in 1 of 18 healthy samples and 38 of 58 disease samples. These data suggest that this phylotype, and the TM7 bacterial division in general, may play a role in the multifactorial process leading to periodontitis.
Figures
FIG. 1.
Phylogeny of representative 16S rDNA sequences of oral TM7 phylotypes. The tree was created by using a maximum-likelihood algorithm and 1,076 positions. The 16S rDNA sequences of S. putrefaciens and E. coli were used as out-groups to root the tree. Phylotypes detected in this study are named with the prefix SBG (for subgingival). The two sequences highlighted in gray are novel phylotypes detected in this study. Bootstrap values greater than 50 are shown. The bar represents evolutionary distance. Accession numbers are given in parentheses.
FIG. 2.
FISH images of TM7 and I025 bacteria. Panel A shows cells staining with YO-PRO-1 iodide (nonspecific nucleic acid dye), panel B shows cells in the same field that hybridize with the TM7 bacterial division probe (labeled with Cy5), and panel C shows a cell that hybridizes with the I025 subdivision-specific probe (labeled with Cy3).
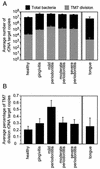
FIG. 3.
Quantification of rDNA from Bacteria and TM7 division members. (A) Average number of bacterial and TM7-division rDNA target copies detected in subgingival crevices with various disease states measured by real-time PCR. (B) Average number of TM7 rDNA target copies as a percentage of the total bacterial rDNA copy number at healthy and diseased sites. Mild-periodontitis sites have significantly higher proportions of TM7 than healthy or severe-periodontitis sites (P < 0.001 and 0.005, respectively). SEs are indicated.
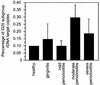
FIG. 4.
Abundance of I025 rDNA relative to total bacterial rDNA in subgingival plaque at sites with various states of disease. Only sites with detectable quantities of I025 are included. SEs are indicated. The difference between relative abundance at sites with mild and moderate periodontitis was significant at P < 0.04.
References
- Chen, R. W., H. Piiparinen, M. Seppänen, P. Koskela, S. Sarna, and M. Lappalainen. 2001. Real-time PCR for detection and quantitation of hepatitis B virus DNA. J. Med. Virol. 65:250-256. - PubMed
- DeSoete, G. 1983. A least squares algorithm for fitting additive trees to proximity data. Psychometrika 48:621-626.
- Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK 56339/DK/NIDDK NIH HHS/United States
- 5T32 AI 07328/AI/NIAID NIH HHS/United States
- T32 AI007328/AI/NIAID NIH HHS/United States
- P60 DE013058/DE/NIDCR NIH HHS/United States
- P60 DE 13058/DE/NIDCR NIH HHS/United States
- P30 DK056339/DK/NIDDK NIH HHS/United States
- R01 DE013541/DE/NIDCR NIH HHS/United States
- R01 DE 13541/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases