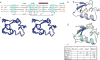Structural insights into the U-box, a domain associated with multi-ubiquitination - PubMed (original) (raw)
Comparative Study
Structural insights into the U-box, a domain associated with multi-ubiquitination
Melanie D Ohi et al. Nat Struct Biol. 2003 Apr.
Abstract
The structure of the U-box in the essential Saccharomyces cerevisiae pre-mRNA splicing factor Prp19p has been determined by NMR. The conserved zinc-binding sites supporting the cross-brace arrangement in RING-finger domains are replaced by hydrogen-bonding networks in the U-box. These hydrogen-bonding networks are necessary for the structural stabilization and activity of the U-box. A conservative Val-->Ile point mutation in the Prp19p U-box domain leads to pre-mRNA splicing defects in vivo. NMR analysis of this mutant shows that the substitution disrupts structural integrity of the U-box domain. Furthermore, comparison of the Prp19p U-box domain with known RING-E2 complex structures demonstrates that both U-box and RING-fingers contain a conserved interaction surface. Mutagenesis of residues at this interface, while not perturbing the structure of the U-box, abrogates Prp19p function in vivo. These comparative structural and functional analyses imply that the U-box and its associated ubiquitin ligase activity are critical for Prp19p function in vivo.
Conflict of interest statement
Competing interests statement The authors declare that they have no competing financial interests.
Figures
Fig. 1
The U-box and RING finger domains share a conserved fold. a, Alignment of selected U-box and RING finger domains. The shadings are yellow for hydrophobic, green for residues important in c-Cbl interaction with its E2 and blue for Zn2+-chelating residues in the RING fingers. Hs represents human proteins; Sc, S. cerevisiae. Blue arrows above the alignment indicate β-strands, and the red cylinder indicates the central α-helix. b, Ribbon diagram of the three-dimensional structure of the Prp19p U-box domain, with core hydrophobic residues in yellow. c, Stereo view of the Cα trace of the 20 lowest energy NMR structures. d, Overlay of the structures of the Prp19p U-box (green) and the c-Cbl RING finger (pink) (PDB entry 1FBV). e, Backbone r.m.s. deviations between the Prp19p U-box and three RING fingers using ProSup. Panels (b_–_d) were generated using MolMol.
Fig. 2
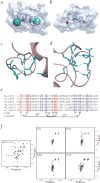
U-box stability is mediated by hydrogen-bonding networks. a, Structure of the Prp19p U-box with the hydrogen-bonding networks represented by a green sphere with a radius of 2.7 Å. Sphere centers are located at the sulfhydryl of Cys3 and carboxylate of Asp38. b, Structure of the c-CBL RING finger with zinc ions represented by a red sphere with a van der Waals radius of 1.3 Å. c,d, Residues involved in the first and second hydrogen-bonding networks. Shown is an optimized close-up view of the two networks in the Prp19p U-box, with donor-acceptor distances ≤2.7 Å highlighted by green dotted lines. Residues selected for subsequent mutagenesis are displayed in green. e, Sequence alignment of several U-box proteins. The eight positions of the zinc ligands in RING proteins are indicated below. Residues structurally identified as participating in the two hydrogen-bonding networks are shown in red and blue. f, 15N-1H HSQC spectra of Prp19p(1–73) and the C3A, E24A, D38A and T41A mutants.
Fig. 3
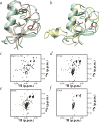
Mutational analysis of putative E2-interacting residues in the U-box. a, Overlay of the three-dimensional structures of the Prp19p U-box (green) and the c-Cbl RING finger (pink) (PDB entry 1FBV). Residues found in the shallow hydrophobic groove are indicated in green for the U-box and in red for c-Cbl. b, Overlay of the three-dimensional structures of the Prp19p U-box (green) and the Rbx1 E3-type RING finger (yellow) (PDB entry 1LDJ). Residues found in the shallow hydrophobic groove are shown in green for the U-box and in red for Rbx1. 15N-1H HSQC spectra of c, Prp19p(1–73) and the d, Y31A; e, D34A; and f, P39A mutants.
Fig. 4
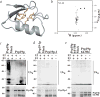
The U-box domain is required for Prp19p function. a, Ribbon diagram of the U-box structure. The Val14 side chain is red, and the side chains of Phe23, Leu28, Ile40, Ile47 and Ile50 are yellow. Val14 has a central role in the protein core, forming extensive contacts with Phe23 and Leu28 in the hydrophobic core, as well as with the aliphatic portions of Lys25 and Glu52, which are solvent exposed. b, 15N-1H HSQC spectrum of Prp19-1p(1–73). Ubiquitination assays preformed with in vitro transcribed and translated c, Prp19p; d, Prp19-1p; or e, Prp19p(64–504). Controls lacking recombinant E1, E2 (Ubc3) and/or Prp19 proteins are indicated in each panel with a minus sign. The reaction mixtures were resolved under reducing conditions by SDS-PAGE, and the separated proteins were subjected to immunoblot analysis with antibodies to ubiquitin (upper panels) or visualized directly by autoradiography (lower panels).
Similar articles
- Structural and functional analysis of essential pre-mRNA splicing factor Prp19p.
Ohi MD, Vander Kooi CW, Rosenberg JA, Ren L, Hirsch JP, Chazin WJ, Walz T, Gould KL. Ohi MD, et al. Mol Cell Biol. 2005 Jan;25(1):451-60. doi: 10.1128/MCB.25.1.451-460.2005. Mol Cell Biol. 2005. PMID: 15601865 Free PMC article. - Structure and biochemical function of a prototypical Arabidopsis U-box domain.
Andersen P, Kragelund BB, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K. Andersen P, et al. J Biol Chem. 2004 Sep 17;279(38):40053-61. doi: 10.1074/jbc.M405057200. Epub 2004 Jun 30. J Biol Chem. 2004. PMID: 15231834 - Characterization of interactions among the Cef1p-Prp19p-associated splicing complex.
Ohi MD, Gould KL. Ohi MD, et al. RNA. 2002 Jun;8(6):798-815. doi: 10.1017/s1355838202025050. RNA. 2002. PMID: 12088152 Free PMC article. - Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing.
Tsai WY, Chow YT, Chen HR, Huang KT, Hong RI, Jan SP, Kuo NY, Tsao TY, Chen CH, Cheng SC. Tsai WY, et al. J Biol Chem. 1999 Apr 2;274(14):9455-62. doi: 10.1074/jbc.274.14.9455. J Biol Chem. 1999. PMID: 10092627 - A new gun in town: the U box is a ubiquitin ligase domain.
Patterson C. Patterson C. Sci STKE. 2002 Jan 22;2002(116):pe4. doi: 10.1126/stke.2002.116.pe4. Sci STKE. 2002. PMID: 11805346 Review.
Cited by
- A ubiquitin-specific, proximity-based labeling approach for the identification of ubiquitin ligase substrates.
Mukhopadhyay U, Levantovsky S, Carusone TM, Gharbi S, Stein F, Behrends C, Bhogaraju S. Mukhopadhyay U, et al. Sci Adv. 2024 Aug 9;10(32):eadp3000. doi: 10.1126/sciadv.adp3000. Epub 2024 Aug 9. Sci Adv. 2024. PMID: 39121224 Free PMC article. - UBE2A and UBE2B are recruited by an atypical E3 ligase module in UBR4.
Barnsby-Greer L, Mabbitt PD, Dery MA, Squair DR, Wood NT, Lamoliatte F, Lange SM, Virdee S. Barnsby-Greer L, et al. Nat Struct Mol Biol. 2024 Feb;31(2):351-363. doi: 10.1038/s41594-023-01192-4. Epub 2024 Jan 5. Nat Struct Mol Biol. 2024. PMID: 38182926 Free PMC article. - Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b.
Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Buchwald G, et al. EMBO J. 2006 Jun 7;25(11):2465-74. doi: 10.1038/sj.emboj.7601144. Epub 2006 May 18. EMBO J. 2006. PMID: 16710298 Free PMC article. - The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing.
Hogg R, McGrail JC, O'Keefe RT. Hogg R, et al. Biochem Soc Trans. 2010 Aug;38(4):1110-5. doi: 10.1042/BST0381110. Biochem Soc Trans. 2010. PMID: 20659013 Free PMC article. Review. - PRP-19, a conserved pre-mRNA processing factor and E3 ubiquitin ligase, inhibits the nuclear accumulation of GLP-1/Notch intracellular domain.
Gutnik S, Thomas Y, Guo Y, Stoecklin J, Neagu A, Pintard L, Merlet J, Ciosk R. Gutnik S, et al. Biol Open. 2018 Jul 16;7(7):bio034066. doi: 10.1242/bio.034066. Biol Open. 2018. PMID: 30012553 Free PMC article.
References
- Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. - PubMed
- Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002;27:368–375. - PubMed
- Patterson C. A new gun in town: the U box is a ubiquitin ligase domain. Sci. STKE. 2002 [online]< http://stke.sciencemag.org/cgi/content/full/OC_sigtraus;2002/116/pe4>. - PubMed
- Koegl M, et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. - PubMed
- Aravind L, Koonin EV. The U box is a modified RING finger — a common domain in ubiquitination. Curr. Biol. 2000;10:R132–R134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases