Effects of the cowpea chlorotic mottle bromovirus beta-hexamer structure on virion assembly - PubMed (original) (raw)
Effects of the cowpea chlorotic mottle bromovirus beta-hexamer structure on virion assembly
D Willits et al. Virology. 2003.
Abstract
The X-ray crystal structure of Cowpea chlorotic mottle bromovirus (CCMV) revealed a unique tubular structure formed by the interaction of the N-termini from six coat protein subunits at each three-fold axis of the assembled virion. This structure, termed the beta-hexamer, consists of six short beta-strands. The beta-hexamer was postulated to play a critical role in the assembly and stability of the virion by stabilizing hexameric capsomers. Mutational analyses of the beta-hexamer structure, utilizing both in vitro and in vivo assembly assays, demonstrate that this structure is not required for virion formation devoid of nucleic acids in vitro or for RNA-containing virions in vivo. However, the beta-hexamer structure does contribute to virion stability in vitro and modulates disease expression in vivo. These results support a model for CCMV assembly through pentamer intermediates.
Figures
Fig. 1
Structural features of CCMV coat protein and virus particles. (A) Ribbon diagram of the CCMV coat protein. The β-strand involved in the β-hexamer formation (amino acids 29–33) is labeled. (B) Enlarged views of a hexamer viewed down the three-fold axis with the β-hexamer emphasized. (C) Side view of the β-hexamer motif.
Fig. 2
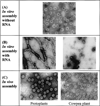
Electron micrographs of CCMV β-hexamer deletion mutants assembled under in vitro and in vivo conditions. (A) Purified NΔβ27–35 coat protein in vitro assembled into T = 3 particles in the absence of viral RNA. (B) Purified NΔβ327–35 coat protein in vitro assembled in the presence of viral RNA showing aberrant assembly products. (C) β-hexdel T = 3 particles purified from protoplasts or plants.
Fig. 3
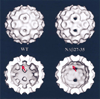
(Top) Shaded-surface representations of the image reconstructions of in vitro assembled CCMV wild-type (WT) and NΔβ27–35 empty particles. (Bottom) Same as (Top) but with front half of particles removed to show the particle interiors indicating the presence (WT) or absence (NΔβ27–35) of β-hexamer density at the pseudo-six-fold axes (arrow).
Fig. 4
Typical symptom expression of cowpea infected with the β-hexdel mutant virus as compared to wild-type virus.
Similar articles
- In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro-transcribed viral cDNA.
Zhao X, Fox JM, Olson NH, Baker TS, Young MJ. Zhao X, et al. Virology. 1995 Mar 10;207(2):486-94. doi: 10.1006/viro.1995.1108. Virology. 1995. PMID: 7886952 - Packaging and structural phenotype of brome mosaic virus capsid protein with altered N-terminal β-hexamer structure.
de Wispelaere M, Chaturvedi S, Wilkens S, Rao AL. de Wispelaere M, et al. Virology. 2011 Oct 10;419(1):17-23. doi: 10.1016/j.virol.2011.07.016. Epub 2011 Aug 23. Virology. 2011. PMID: 21864876 - Comparison of the native CCMV virion with in vitro assembled CCMV virions by cryoelectron microscopy and image reconstruction.
Fox JM, Wang G, Speir JA, Olson NH, Johnson JE, Baker TS, Young MJ. Fox JM, et al. Virology. 1998 Apr 25;244(1):212-8. doi: 10.1006/viro.1998.9107. Virology. 1998. PMID: 9581792 - Quasi-equivalent viruses: a paradigm for protein assemblies.
Johnson JE, Speir JA. Johnson JE, et al. J Mol Biol. 1997 Jun 27;269(5):665-75. doi: 10.1006/jmbi.1997.1068. J Mol Biol. 1997. PMID: 9223631 Review. - Microsecond time-resolved cryo-electron microscopy.
Lorenz UJ. Lorenz UJ. Curr Opin Struct Biol. 2024 Aug;87:102840. doi: 10.1016/j.sbi.2024.102840. Epub 2024 May 28. Curr Opin Struct Biol. 2024. PMID: 38810313 Review.
Cited by
- Controlling viral capsid assembly with templating.
Hagan MF. Hagan MF. Phys Rev E Stat Nonlin Soft Matter Phys. 2008 May;77(5 Pt 1):051904. doi: 10.1103/PhysRevE.77.051904. Epub 2008 May 8. Phys Rev E Stat Nonlin Soft Matter Phys. 2008. PMID: 18643099 Free PMC article. - Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Kryshtafovych A, Moult J, Bales P, Bazan JF, Biasini M, Burgin A, Chen C, Cochran FV, Craig TK, Das R, Fass D, Garcia-Doval C, Herzberg O, Lorimer D, Luecke H, Ma X, Nelson DC, van Raaij MJ, Rohwer F, Segall A, Seguritan V, Zeth K, Schwede T. Kryshtafovych A, et al. Proteins. 2014 Feb;82 Suppl 2(0 2):26-42. doi: 10.1002/prot.24489. Proteins. 2014. PMID: 24318984 Free PMC article. - Mechanisms of size control and polymorphism in viral capsid assembly.
Elrad OM, Hagan MF. Elrad OM, et al. Nano Lett. 2008 Nov;8(11):3850-7. doi: 10.1021/nl802269a. Epub 2008 Oct 25. Nano Lett. 2008. PMID: 18950240 Free PMC article. - Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
Speir JA, Bothner B, Qu C, Willits DA, Young MJ, Johnson JE. Speir JA, et al. J Virol. 2006 Apr;80(7):3582-91. doi: 10.1128/JVI.80.7.3582-3591.2006. J Virol. 2006. PMID: 16537626 Free PMC article.
References
- Adolph KW, Butler PJG. Studies on the assembly of a spherical plant virus. I. States of aggregation of the isolated protein. J. Mol. Biol. 1974;88:327–341. - PubMed
- Ahlquist P. Bromovirus RNA replication and transcription. Curr. Opin. Gen. and Dev. 1992;2(1):71–76. - PubMed
- Bancroft JB, Bracker CE, Wagner GW. Structures derived from cowpea chlorotic mottle and brome mosaic virus protein. Virology. 1969;38:324–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources