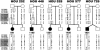Genome architecture catalyzes nonrecurrent chromosomal rearrangements - PubMed (original) (raw)
. 2003 May;72(5):1101-16.
doi: 10.1086/374385. Epub 2003 Mar 20.
Affiliations
- PMID: 12649807
- PMCID: PMC1180264
- DOI: 10.1086/374385
Genome architecture catalyzes nonrecurrent chromosomal rearrangements
Paweł Stankiewicz et al. Am J Hum Genet. 2003 May.
Abstract
To investigate the potential involvement of genome architecture in nonrecurrent chromosome rearrangements, we analyzed the breakpoints of eight translocations and 18 unusual-sized deletions involving human proximal 17p. Surprisingly, we found that many deletion breakpoints occurred in low-copy repeats (LCRs); 13 were associated with novel large LCR17p structures, and 2 mapped within an LCR sequence (middle SMS-REP) within the Smith-Magenis syndrome (SMS) common deletion. Three translocation breakpoints involving 17p11 were found to be located within the centromeric alpha-satellite sequence D17Z1, three within a pericentromeric segment, and one at the distal SMS-REP. Remarkably, our analysis reveals that LCRs constitute >23% of the analyzed genome sequence in proximal 17p--an experimental observation two- to fourfold higher than predictions based on virtual analysis of the genome. Our data demonstrate that higher-order genomic architecture involving LCRs plays a significant role not only in recurrent chromosome rearrangements but also in translocations and unusual-sized deletions involving 17p.
Figures
Figure 1
Breakpoint analysis of unusual-sized deletions. Proximal chromosome 17p is depicted at the bottom, showing the size, position, and orientation of LCRs. Dashed horizontal lines represent the genomic segment deleted for 18 different patients, and solid horizontal lines depict the retained genomic material, with the patient number shown to the right. The LCR17p structures are depicted in colors, to better represent their homology and orientation with respect to each other; the closed arrowheads represent the orientation of the LCR17p subunits. Selected breakpoints involving the LCR17ps are shown as vertical dashed lines. The horizontal line flanked by open arrowheads (below the genomic segments) depicts the SMS critical region; the common deletion (80%–90% of patients with SMS) occurs between proximal and distal SMS-REP copies. Note that the distal deletion breakpoints in patients 357, 993, and 2011 map outside the analyzed genomic region and thus were not included in the calculation of the percentage of chromosome breakpoints associated with LCRs in proximal 17p. Only >20-kb LCRs are depicted. The map is not to scale.
Figure 2
Schematic representation of a dual-color interphase FISH assay developed to screen for common, A, versus unusual-sized SMS deletions. The map of chromosome 17p11.2 with the placement of the FISH probes for one chromosome homologue is shown at the top of the figure. The proximal SMS-REP–flanking clones BAC RP11-344E13 and PAC RP5-836L9 and the distal SMS-REP flanking BACs RP11-416I2 and RP11-209J20 are differentially labeled and are detected with red and green colors, respectively. Below the chromosome map, in the left chamber of the slide, two adjacent green and red dots represent the normal chromosome 17, and the presence of a single green signal demonstrates that the deletion breakpoint occurred between clones RP11-209J20 and RP11-416I2, within the distal SMS-REP. Similarly, the absence of the second green signal on the right side indicates that the breakpoint maps between clones RP5-836L9 and RP11-344E13. The red and green signals flanking SMS-REPs do not overlap, because the distance between the clones is greater than the ∼100-kb resolution limit of interphase FISH. The three other hypothetical microscope slides give examples of the FISH results obtained with the same clones. B, A small deletion with the telomeric breakpoint mapping within the distal SMS-REP and the centromeric breakpoint mapping between the proximal and distal SMS-REPs. C, A large deletion with the distal breakpoint mapping telomeric to the distal SMS-REP and the centromeric breakpoint mapping within the proximal SMS-REP. D, A large deletion with the telomeric breakpoint mapping within the distal SMS-REP and the proximal breakpoint mapping centromeric to the proximal SMS-REP.
Figure 3
Summary of deletion breakpoint mapping. A, Schematic representation of LCRs within 17p11.2-p12, with horizontal lines attached to the table showing the position of individual BAC/PAC clones. B, A table with the clones used in FISH studies. PAC/BAC clones that gave positive FISH signals are represented by filled blue bars, and white spaces depict BAC/PAC clones that did not give a hybridization signal from the deleted chromosome (however, each individual clone was not assayed by FISH). Clones, within which the breakpoint was mapped, are depicted by half blue-green shading. The gray vertical shading indicates the three SMS-REPs. Selected clones used for the FISH analysis of uncommon deletions are taken from the complete BAC/PAC contig (Bi et al. 2002) and are labeled in the upper row of the table. Our analysis of the currently available databases reveals that clones RP11-416I2 and RP11-45M22 appear not to overlap and are spanned by the BAC clone RP11-367G9.
Figure 4
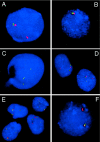
FISH analyses of interphase nuclei used to map the rearrangement breakpoints. A and B, Interphase nuclei of patient 1939 after FISH with SMS-REPs–flanking clones (fig. 2). In A, the absence of the green signal (RP5-836L9) and presence of the red signal (RP11-344E13) on the del(17) indicates that the proximal breakpoint maps within the proximal SMS-REP (or directly adjacent LCR17pB). In B, the absence of both red (RP11-416I2) and green (RP11-209J20) signals on the del(17) shows that the distal breakpoint maps telomeric to the distal SMS-REP. C, FISH with BAC clone CIT-3157E16 (LCR17pA), enabling mapping the distal breakpoint of the deletion in patient 572 to the distal portion of LCR17pA. The two closely spaced green signals on the normal and deleted chromosomes 17 represent LCR17pC and LCR17pD copies. A single hybridization signal corresponds to the LCR17pA copy on the normal chromosome 17; the LCR17pA on der(17) is deleted. D, FISH with the BAC RP11-344E13 (red) and a centromeric probe (green) on cells from patient GM02836, showing the breakpoint mapped between them. On the normal chromosome 17, the red and green signals are relatively close to each other, whereas the separation of the red and green signals indicates that they are on different chromosomes, der(9) and der(17), respectively. E, The centromeric breakpoint on chromosome 17 in the patient GM03119, identified after the cohybridization of BAC RP11-344E13 (red) and the centromeric probe D17Z1 (green). In addition to the adjacent red and green pair of signals on both chromosomes 17 and the der(9), the single green signal on the der(17) is of weaker intensity when compared with the other two green signals, indicating the localization of this breakpoint to the q11.1 portion of the chromosome 17 centromere. Note the variability of distances between RP11-344E13 (red) and the D17Z1 centromeric probe (green) in D and E, demonstrating different condensation of the pericentromeric heterochromatin. F, FISH with the distal SMS-REP flanking BAC clones RP11-209J20 (green) and RP11-416I2 (red) on an interphase nucleus of patient 1576, a carrier of an unbalanced translocation. The presence of only the red signal on the der(17) chromosome indicates that the breakpoint maps between these two clones.
Figure 5
A, PFGE detection of novel junction fragments in three patients with SMS (patients 540, 641, and 1456) with smaller-sized deletions in which the proximal breakpoint maps to middle SMS-REP. The Southern blot was hybridized with a PRPSAP2 PCR probe, which, in addition to the normal 1.4-Mb _Not_I fragment (control patients 642 and 644), identified the 0.9-, 1.1-, and 1.7-Mb junction fragments (arrows) spanning the proximal breakpoints within middle SMS-REP or LCR17pB. B, Schematic diagram represents the derivation of novel PFGE junction fragments in patients with SMS with uncommon deletions.
Figure 6
Haplotypes of five patients with unusual deletions and their families. Standard pedigree symbols are used; a circle denotes a female, a square denotes a male. Blackened circles or squares indicate an affected individual. To the left of each pedigree is a list of microsatellite markers used for genotyping; those within the SMS common deletion region are bold and shaded. The allele numbers are located under each family member. The genotypes of markers within the SMS common deletion region are bold in the patients and the parent of origin. The dotted lines outline alleles inherited by the patient from the parent of origin. In patient 641, recombination occurred between the region flanked by loci D17S122 and D17S1857 and the region between D17S2257 and D17S805 (including the middle SMS-REP), resulting in the deletion. In patient 1190, recombination between the region flanked by markers D17S1857 and D17S2258 (including the distal SMS-REP) and the region flanked by markers D17S2257 and D17S805 (including the middle SMS-REP) resulted in the deletion. Recombination between the region flanked by loci D17S1857 and D17S2258 and the region between D17S2259 and D17S842 (including the proximal SMS-REP) resulted in the deletion in patient 1354. Patients 1456 and 1931 may have deletions resulting from intrachromosomal recombination. (Both of these patients had crossovers on their intact, maternally derived chromosomes 17 [between loci D17S842 and D17S1871 for patient 1456 and between loci D17S955 and D17S122 for patient 1931]). Interestingly, each of the five deletions are paternally derived, as evidenced by the lack of a paternal allele for loci D17S1857, D17S2258, D17S2256, and D17S2257 for patient 641; loci D17S2258, D17S2256, and D17S2257 for patients 1190 and 1456; and loci D17S2258, D17S2256, D17S2257, D17S805, and D17S2259 for patients 1354 and 1931. The locations of markers used in genotyping are shown in figure 1.
Figure 7
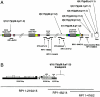
A, Schematic diagram of identified translocation breakpoints within proximal 17p. FISH experiments on cells from patients harboring translocations with breakpoints in proximal 17p (table 1) showed that five of eight analyzed breakpoints cluster centromeric to the most proximal BAC clone RP11-728E14: one within clone RP11-344E13, one within two overlapping BAC clones (CTD-2354J3 and RP11-311F12), and one at the centromeric end of the distal SMS-REP, within the PAC clone RP1-48J12. B, Schematic representation of the chromosome 17 translocation breakpoint in patient 1576. The breakpoint was mapped between the BAC clones RP11-416I2 and RP11-209J20, indicating that it occurred within or adjacent to the distal SMS-REP on the centromeric side. FISH with the long-range PCR product specific to the KER gene cluster localized within the ∼10–42-kb proximal portion of the distal SMS-REP (Park et al. 2002) showed that it was translocated on the der(10) chromosome. Subsequent FISH mapping with the PAC clone RP1-48J12 that overlaps the distal SMS-REP by ∼20 kb showed that only a small fragment of the clone RP1-48J2 was translocated. Thus, the chromosome 17 breakpoint was mapped at the proximal end of the distal SMS-REP.
Similar articles
- Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders.
Shaw CJ, Shaw CA, Yu W, Stankiewicz P, White LD, Beaudet AL, Lupski JR. Shaw CJ, et al. J Med Genet. 2004 Feb;41(2):113-9. doi: 10.1136/jmg.2003.012831. J Med Genet. 2004. PMID: 14757858 Free PMC article. - Complex chromosome 17p rearrangements associated with low-copy repeats in two patients with congenital anomalies.
Vissers LE, Stankiewicz P, Yatsenko SA, Crawford E, Creswick H, Proud VK, de Vries BB, Pfundt R, Marcelis CL, Zackowski J, Bi W, van Kessel AG, Lupski JR, Veltman JA. Vissers LE, et al. Hum Genet. 2007 Jul;121(6):697-709. doi: 10.1007/s00439-007-0359-6. Epub 2007 Apr 25. Hum Genet. 2007. PMID: 17457615 Free PMC article. - Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs.
Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR. Park SS, et al. Genome Res. 2002 May;12(5):729-38. doi: 10.1101/gr.82802. Genome Res. 2002. PMID: 11997339 Free PMC article. - Trisomy 17p10-p12 due to mosaic supernumerary marker chromosome: delineation of molecular breakpoints and clinical phenotype, and comparison to other proximal 17p segmental duplications.
Yatsenko SA, Treadwell-Deering D, Krull K, Lewis RA, Glaze D, Stankiewicz P, Lupski JR, Potocki L. Yatsenko SA, et al. Am J Med Genet A. 2005 Oct 1;138A(2):175-80. doi: 10.1002/ajmg.a.30948. Am J Med Genet A. 2005. PMID: 16152635 Review. - Jumping translocations.
Berger R, Bernard OA. Berger R, et al. Genes Chromosomes Cancer. 2007 Aug;46(8):717-23. doi: 10.1002/gcc.20456. Genes Chromosomes Cancer. 2007. PMID: 17444494 Review.
Cited by
- Breakpoint mapping and haplotype analysis of three reciprocal translocations identify a novel recurrent translocation in two unrelated families: t(4;11)(p16.2;p15.4).
Thomas NS, Maloney V, Bryant V, Huang S, Brewer C, Lachlan K, Jacobs PA. Thomas NS, et al. Hum Genet. 2009 Mar;125(2):181-8. doi: 10.1007/s00439-008-0611-8. Epub 2008 Dec 24. Hum Genet. 2009. PMID: 19104840 - Copy number variation at the breakpoint region of isochromosome 17q.
Carvalho CM, Lupski JR. Carvalho CM, et al. Genome Res. 2008 Nov;18(11):1724-32. doi: 10.1101/gr.080697.108. Epub 2008 Aug 19. Genome Res. 2008. PMID: 18714090 Free PMC article. - Neurodevelopmental Genetic Diseases Associated With Microdeletions and Microduplications of Chromosome 17p13.3.
Blazejewski SM, Bennison SA, Smith TH, Toyo-Oka K. Blazejewski SM, et al. Front Genet. 2018 Mar 23;9:80. doi: 10.3389/fgene.2018.00080. eCollection 2018. Front Genet. 2018. PMID: 29628935 Free PMC article. Review. - Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements.
Froyen G, Belet S, Martinez F, Santos-Rebouças CB, Declercq M, Verbeeck J, Donckers L, Berland S, Mayo S, Rosello M, Pimentel MM, Fintelman-Rodrigues N, Hovland R, Rodrigues dos Santos S, Raymond FL, Bose T, Corbett MA, Sheffield L, van Ravenswaaij-Arts CM, Dijkhuizen T, Coutton C, Satre V, Siu V, Marynen P. Froyen G, et al. Am J Hum Genet. 2012 Aug 10;91(2):252-64. doi: 10.1016/j.ajhg.2012.06.010. Epub 2012 Jul 26. Am J Hum Genet. 2012. PMID: 22840365 Free PMC article. - Rational design and construction of multi-copy biomanufacturing islands in mammalian cells.
Altamura R, Doshi J, Benenson Y. Altamura R, et al. Nucleic Acids Res. 2022 Jan 11;50(1):561-578. doi: 10.1093/nar/gkab1214. Nucleic Acids Res. 2022. PMID: 34893882 Free PMC article.
References
Electronic-Database Information
- BACPAC Resources Center Home Page, Children’s Hospital Oakland, http://www.chori.org/bacpac/
- Coriell Cell Repositories, http://locus.umdnj.edu/
- NCBI BLAST Home Page, http://www.ncbi.nlm.nih.gov/blast/
- NCBI Home Page, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMS) - PubMed
References
- Bandyopadhyay R, Berend SA, Page SL, Choo KHA, Shaffer LG (2001) Satellite III sequences on 14p and their relevance to Robertsonian translocation formation. Chromosome Res 9:235–242 - PubMed
- Beheshti B, Karaskova J, Park PC, Squire JA, Beatty BG (2000) Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer cell lines by sequential Giemsa banding and spectral karyotyping. Mol Diagn 5:23–32 - PubMed
- Berger R, Busson-Le Coniat M (1999) Centric and pericentric chromosome rearrangements in hematopoietic malignancies. Leukemia 13:671–678 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials