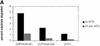Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate - PubMed (original) (raw)
Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate
Mingsheng Zhang et al. EMBO J. 2003.
Abstract
Ornithine decarboxylase (ODC) is regulated by its metabolic products through a feedback loop that employs a second protein, antizyme 1 (AZ1). AZ1 accelerates the degradation of ODC by the proteasome. We used purified components to study the structural elements required for proteasomal recognition of this ubiquitin-independent substrate. Our results demonstrate that AZ1 acts on ODC to enhance the association of ODC with the proteasome, not the rate of its processing. Substrate-linked or free polyubiquitin chains compete for AZ1-stimulated degradation of ODC. ODC-AZ1 is therefore recognized by the same element(s) in the proteasome that mediate recognition of polyubiquitin chains. The 37 C-terminal amino acids of ODC harbor an AZ1-modulated recognition determinant. Within the ODC C terminus, three subsites are functionally distinguishable. The five terminal amino acids (ARINV, residues 457-461) collaborate with residue C441 to constitute one recognition element, and AZ1 collaborates with additional constituents of the ODC C terminus to generate a second recognition element.
Figures
Fig. 1. ODC degradation by the 26S proteasome in vitro. The extent of degradation was determined after 30 min in reaction mixtures with 50 nM proteasomes, 50 nM ODC and in the presence or absence of 400 nM AZ1. The effects of the proteasome inhibitors MG132 and epoxomicin and of ATP depletion were tested.
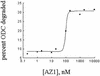
Fig. 2. Dependence of ODC degradation on AZ1 concentration. The extent of degradation was determined after 50 min in reaction mixtures with 50 nM proteasomes, 50 nM ODC and the indicated concentrations of AZ1.
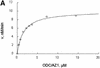
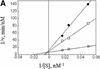
Fig. 3. AZ1 increases the apparent affinity of ODC but not the rate of catalysis. Incubations were carried out for 30 min and contained 50 nM 26S proteasomes (A and B) or 100 nM proteasomes (C and D), 50 nM [35S]ODC and various amounts of total ODC (labeled plus unlabeled), as indicated. (A and B) Reactions with 12 µM AZ1. (C and D) Reactions with no AZ1 present. (A and C) Velocity of degradation versus substrate concentration. The curves are a least-squares fit of the Michaelis–Menten equation assuming a _K_m of 1.6 µM (A) or of 13 µM (C). (B and D) Double reciprocal (Lineweaver–Burk) plots of data in (A and C), respectively.
Fig. 3. AZ1 increases the apparent affinity of ODC but not the rate of catalysis. Incubations were carried out for 30 min and contained 50 nM 26S proteasomes (A and B) or 100 nM proteasomes (C and D), 50 nM [35S]ODC and various amounts of total ODC (labeled plus unlabeled), as indicated. (A and B) Reactions with 12 µM AZ1. (C and D) Reactions with no AZ1 present. (A and C) Velocity of degradation versus substrate concentration. The curves are a least-squares fit of the Michaelis–Menten equation assuming a _K_m of 1.6 µM (A) or of 13 µM (C). (B and D) Double reciprocal (Lineweaver–Burk) plots of data in (A and C), respectively.
Fig. 3. AZ1 increases the apparent affinity of ODC but not the rate of catalysis. Incubations were carried out for 30 min and contained 50 nM 26S proteasomes (A and B) or 100 nM proteasomes (C and D), 50 nM [35S]ODC and various amounts of total ODC (labeled plus unlabeled), as indicated. (A and B) Reactions with 12 µM AZ1. (C and D) Reactions with no AZ1 present. (A and C) Velocity of degradation versus substrate concentration. The curves are a least-squares fit of the Michaelis–Menten equation assuming a _K_m of 1.6 µM (A) or of 13 µM (C). (B and D) Double reciprocal (Lineweaver–Burk) plots of data in (A and C), respectively.
Fig. 3. AZ1 increases the apparent affinity of ODC but not the rate of catalysis. Incubations were carried out for 30 min and contained 50 nM 26S proteasomes (A and B) or 100 nM proteasomes (C and D), 50 nM [35S]ODC and various amounts of total ODC (labeled plus unlabeled), as indicated. (A and B) Reactions with 12 µM AZ1. (C and D) Reactions with no AZ1 present. (A and C) Velocity of degradation versus substrate concentration. The curves are a least-squares fit of the Michaelis–Menten equation assuming a _K_m of 1.6 µM (A) or of 13 µM (C). (B and D) Double reciprocal (Lineweaver–Burk) plots of data in (A and C), respectively.
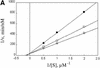
Fig. 4. Competition between ODC–AZ1 and Ub5DHFR. (A) Com petitive inhibition of ODC–AZ1 by Ub5DHFR. Incubations contained 50 nM rat 26S proteasomes, 12 µM AZ1 with 0.5, 1 or 2 µM [35S]ODC and 0 (open circles), 100 nM (squares) or 500 nM (filled circles) Ub5DHFR. (B) Competitive inhibition of Ub5DHFR by ODC–AZ1. Incubations contained 2.5 nM rat 26S proteasomes, with 20, 40 or 60 nM [32P]Ub5DHFR, 12 µM AZ1, and 0 (open circles), 2.5 µM (squares) or 5 µM (filled circles) ODC–AZ1.
Fig. 4. Competition between ODC–AZ1 and Ub5DHFR. (A) Com petitive inhibition of ODC–AZ1 by Ub5DHFR. Incubations contained 50 nM rat 26S proteasomes, 12 µM AZ1 with 0.5, 1 or 2 µM [35S]ODC and 0 (open circles), 100 nM (squares) or 500 nM (filled circles) Ub5DHFR. (B) Competitive inhibition of Ub5DHFR by ODC–AZ1. Incubations contained 2.5 nM rat 26S proteasomes, with 20, 40 or 60 nM [32P]Ub5DHFR, 12 µM AZ1, and 0 (open circles), 2.5 µM (squares) or 5 µM (filled circles) ODC–AZ1.
Fig. 5. Competition by Ub4. (A) Competitive inhibition of Ub5DHFR by Ub4. Incubations contained 2.5 nM rat 26S proteasomes, with 20, 40 or 60 nM [32P]Ub5DHFR, and 0 (open circles), 0.8 µM (squares) or 2 µM (filled circles) Ub4. (B) Competitive inhibition of ODC–AZ1 by Ub4. Incubations contained 50 nM rat 26S proteasomes, with 12 µM AZ1 and 0.5, 1 or 2 µM [35S]ODC, and no (open circles), 0.8 µM (squares) or 2 µM (filled circles) Ub4.
Fig. 5. Competition by Ub4. (A) Competitive inhibition of Ub5DHFR by Ub4. Incubations contained 2.5 nM rat 26S proteasomes, with 20, 40 or 60 nM [32P]Ub5DHFR, and 0 (open circles), 0.8 µM (squares) or 2 µM (filled circles) Ub4. (B) Competitive inhibition of ODC–AZ1 by Ub4. Incubations contained 50 nM rat 26S proteasomes, with 12 µM AZ1 and 0.5, 1 or 2 µM [35S]ODC, and no (open circles), 0.8 µM (squares) or 2 µM (filled circles) Ub4.
Fig. 6. Effect of C-terminal truncations and mutation on ODC degradation. The extent of degradation was determined after 30 min in incubations containing 50 nM proteasome, with or without 400 nM AZ1, as indicated.
Fig. 7. Degradation of T.brucei ODC with mouse ODC C-terminal extensions. The extent of degradation was determined after 30 min in incubations containing 100 nM proteasome, with or without 400 nM AZ1, as indicated.
Fig. 8. Inhibition of ODC degradation by GFP with mouse ODC C-terminal extensions. Data are normalized to ODC degradation observed in the absence of competing inhibitor protein and is expressed as percent residual degradation. Incubations were for 30 min and contained 100 nM proteasome, 50 nM ODC and various concentrations of inhibitors, as indicated. The extent of ODC degradation in the absence of inhibitors was 5.4%.
Fig. 9. DHFR with ODC C-terminal extensions: degradation and competitive inhibition. (A) Degradation of DHFR with ODC C-terminal extensions. The extent of degradation was determined after 30 min in incubations containing 100 nM proteasome, with or without 50 µM methotrexate, as indicated. (B) Inhibition of ODC degradation by DHFR with mouse ODC C-terminal extensions. Incubations were performed in the absence of methotrexate (open symbols, ×) or in the presence of 50 µM methotrexate (filled symbols). Conditions of incubation and presentation of data are as in Figure 8. The extent of ODC degradation in the absence of inhibitors was 6.3%.
Fig. 9. DHFR with ODC C-terminal extensions: degradation and competitive inhibition. (A) Degradation of DHFR with ODC C-terminal extensions. The extent of degradation was determined after 30 min in incubations containing 100 nM proteasome, with or without 50 µM methotrexate, as indicated. (B) Inhibition of ODC degradation by DHFR with mouse ODC C-terminal extensions. Incubations were performed in the absence of methotrexate (open symbols, ×) or in the presence of 50 µM methotrexate (filled symbols). Conditions of incubation and presentation of data are as in Figure 8. The extent of ODC degradation in the absence of inhibitors was 6.3%.
Similar articles
- Structural basis of antizyme-mediated regulation of polyamine homeostasis.
Wu HY, Chen SF, Hsieh JY, Chou F, Wang YH, Lin WT, Lee PY, Yu YJ, Lin LY, Lin TS, Lin CL, Liu GY, Tzeng SR, Hung HC, Chan NL. Wu HY, et al. Proc Natl Acad Sci U S A. 2015 Sep 8;112(36):11229-34. doi: 10.1073/pnas.1508187112. Epub 2015 Aug 24. Proc Natl Acad Sci U S A. 2015. PMID: 26305948 Free PMC article. - Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination.
Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Murakami Y, et al. Nature. 1992 Dec 10;360(6404):597-9. doi: 10.1038/360597a0. Nature. 1992. PMID: 1334232 - Structural elements of antizymes 1 and 2 are required for proteasomal degradation of ornithine decarboxylase.
Chen H, MacDonald A, Coffino P. Chen H, et al. J Biol Chem. 2002 Nov 29;277(48):45957-61. doi: 10.1074/jbc.M206799200. Epub 2002 Sep 30. J Biol Chem. 2002. PMID: 12359729 - Turnover of trypanosomal ornithine decarboxylases.
Persson L, Jeppsson A, Nasizadeh S. Persson L, et al. Biochem Soc Trans. 2003 Apr;31(2):411-4. doi: 10.1042/bst0310411. Biochem Soc Trans. 2003. PMID: 12653649 Review. - Degradation of ornithine decarboxylase by the 26S proteasome.
Murakami Y, Matsufuji S, Hayashi S, Tanahashi N, Tanaka K. Murakami Y, et al. Biochem Biophys Res Commun. 2000 Jan 7;267(1):1-6. doi: 10.1006/bbrc.1999.1706. Biochem Biophys Res Commun. 2000. PMID: 10623564 Review.
Cited by
- Mechanisms and regulation of substrate degradation by the 26S proteasome.
Arkinson C, Dong KC, Gee CL, Martin A. Arkinson C, et al. Nat Rev Mol Cell Biol. 2024 Oct 3. doi: 10.1038/s41580-024-00778-0. Online ahead of print. Nat Rev Mol Cell Biol. 2024. PMID: 39362999 Review. - Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Yan X, Yuan X, Lv J, Zhang B, Huang Y, Li Q, Ma J, Li Y, Wang X, Li Y, Yu Y, Liu Q, Liu T, Mi W, Dong C. Yan X, et al. Nat Commun. 2024 Feb 8;15(1):1170. doi: 10.1038/s41467-024-45521-7. Nat Commun. 2024. PMID: 38326322 Free PMC article. - Ubiquitin-proteasome pathway and cellular responses to oxidative stress.
Shang F, Taylor A. Shang F, et al. Free Radic Biol Med. 2011 Jul 1;51(1):5-16. doi: 10.1016/j.freeradbiomed.2011.03.031. Epub 2011 Apr 8. Free Radic Biol Med. 2011. PMID: 21530648 Free PMC article. Review. - Imaging of radiation effects on cellular 26S proteasome function in situ.
Brush JM, Kim K, Sayre JW, McBride WH, Iwamoto KS. Brush JM, et al. Int J Radiat Biol. 2009 Jun;85(6):483-94. doi: 10.1080/09553000902883794. Int J Radiat Biol. 2009. PMID: 19401903 Free PMC article. - Timely activation of budding yeast APCCdh1 involves degradation of its inhibitor, Acm1, by an unconventional proteolytic mechanism.
Melesse M, Choi E, Hall H, Walsh MJ, Geer MA, Hall MC. Melesse M, et al. PLoS One. 2014 Jul 29;9(7):e103517. doi: 10.1371/journal.pone.0103517. eCollection 2014. PLoS One. 2014. PMID: 25072887 Free PMC article.
References
- Almrud J.J., Oliveira,M.A., Kern,A.D., Grishin,N.V., Phillips,M.A. and Hackert,M.L. (2000) Crystal structure of human ornithine decarboxylase at 2.1 Å resolution: structural insights to antizyme binding. J. Mol. Biol., 295, 7–16. - PubMed
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The protea some: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. - PubMed
- Bercovich Z., Rosenberg-Hasson,Y., Ciechanover,A. and Kahana,C. (1989) Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J. Biol. Chem., 264, 15949–15952. - PubMed
- Braun B.C., Glickman,M., Kraft,R., Dahlmann,B., Kloetzel,P.M., Finley,D. and Schmidt,M. (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol., 1, 221–226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials