Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast - PubMed (original) (raw)
Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast
Carine Ganem et al. EMBO J. 2003.
Abstract
Ssu72 is an essential yeast protein that is involved in transcription. It physically interacts with transcription initiation and termination complexes. In this report, we provide evidence that Ssu72 is a phosphatase that physically interacts with the CTD kinase Kin28 and functionally interacts with the CTD phosphatase Fcp1. A genome-wide expression analysis of mutant ssu72-ts69 during growth in complete medium revealed a number of defects, including the accumulation of a limited number of mRNAs and the read-through transcription of small nucleolar RNAs and of some mRNAs. We hypothesize that Ssu72 plays a key role in the transcription termination of certain transcripts, possibly by promoting RNA polymerase pausing and release. The possibility that the CTD of the largest subunit of RNA polymerase II is a substrate of Ssu72 is discussed.
Figures
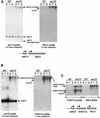
Fig. 1. Read-through transcription of snoRNAs in the ssu72-ts69 mutant. Northern blot analyses of transcripts in wild-type (WT) and ssu72-ts69 (ssu72) cells 0, 1 or 2 h after a shift to 39°C. The probes used are indicated at the bottom of each autoradiography, together with a schematic representation of the relative position of each gene analysed in the yeast genome. The different RNA transcripts are indicated with arrows. The positions of wild-type forms of YCR015c and TRS31 transcripts have been indicated as seen on overexposed autoradiographies (data not shown). (A) Probes for snr13 and TRS31 transcripts. (B) Probes for snr33 and Ycr015c transcripts. (C) Probes for POL4 and YCR015c transcripts.
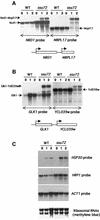
Fig. 2. Accumulation and read-through transcription of mRNAs in the ssu72-ts69 mutant. Northern blot analyses of transcripts in wild-type (WT) and ssu72-ts69 (Ssu72) cells 0, 1 or 2 h after a shift to 39°C. The probes used are indicated at the bottom (A and B) or at the right (C) of each autoradiography. For (A) and (B), a schematic representation of the relative position of each gene analyzed in the yeast genome is available. (A) Accumulation and read-through transcription of the NRD1 mRNA. (B) Read-through transcription of the GLK1 mRNA. (C) Accumulation of HRP1 and HSP30 transcripts. ACT1 probes and ribosomal RNAs are used as invariant controls.
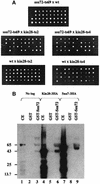
Fig. 3. (A) Assay of synthetic lethality. Strains GF3969 (ssu72-ts69) was crossed with strain GF4040 (kin28-ts2) or GF4046 (kin28-ts4). As controls we crossed these mutants with wild-type strains GF1084 or GF1083. Diploids obtained were induced to sporulate. After dissection, segregants were left growing for 3 days at 28°C on YPD plates. (B) Pull-down experiments. Lanes 1, 4 and 7: 50 µg of yeast crude extracts containing no HA-tagged protein (1), protein Kin28 HA-tagged (4) and protein Sua7 HA-tagged (7) were acetone precipitated then loaded directly on the SDS–PAGE gel. Lanes 2 and 3: 50 µg of yeast crude extract containing no protein HA-tagged were incubated with glutathione–Sepharose beads coated with either GST alone (2) or GST–Ssu72 fusion protein (3), then the complexes were processed as described in Materials and methods. After acetone precipitation, the eluates were loaded on the SDS–PAGE gel. Lanes 5 and 6: 50 µg of yeast crude extract containing protein Kin28 HA-tagged were incubated with either GST alone (5) or GST–Ssu72 fusion protein (6) then processed as above. Lanes 8 and 9: 50 µg of yeast crude extract containing protein Sua7 HA-tagged were incubated with either GST alone (8) or GST–Ssu72 fusion protein (9) then processed as above. HA-tagged proteins were revealed with anti-HA 12CA5 antibody.
Fig. 4. (A) Structural alignment of Ssu72 of S.cerevisiae, eukaryotic orthologues and LMW-PTPs. The figure was made using the program Espript (Gouet et al., 1999). Ssu72: Ssu72 S.cerevisiae; ss72_arath: Ssu72 Arabidopsis thaliana; ss72_schpo: Ssu72 Schizosaccharomyces pombe; ss72_human: Ssu72 Homo sapiens; ss72_drome: Ssu72 Drosophila melanogaster; ss72_enccu: Ssu72 Encephalitozoon cuniculi; 5pnt: human LMW.PTP (HCPTP-A); 1phr: BPTP Bos taurus; 1d1q: LTP1: S.cerevisiae. (B) Tridimensional model of Ssu72 compared with the structure of Ltp1 obtained by X-ray diffraction (Wang et al., 2000b).
Fig. 4. (A) Structural alignment of Ssu72 of S.cerevisiae, eukaryotic orthologues and LMW-PTPs. The figure was made using the program Espript (Gouet et al., 1999). Ssu72: Ssu72 S.cerevisiae; ss72_arath: Ssu72 Arabidopsis thaliana; ss72_schpo: Ssu72 Schizosaccharomyces pombe; ss72_human: Ssu72 Homo sapiens; ss72_drome: Ssu72 Drosophila melanogaster; ss72_enccu: Ssu72 Encephalitozoon cuniculi; 5pnt: human LMW.PTP (HCPTP-A); 1phr: BPTP Bos taurus; 1d1q: LTP1: S.cerevisiae. (B) Tridimensional model of Ssu72 compared with the structure of Ltp1 obtained by X-ray diffraction (Wang et al., 2000b).
Fig. 5. (A) Samples of soluble crude extracts prepared from E.coli cells transformed with plasmids pET21a (1), pET21a-His6-SSU72 (2), pET21a-SSU72-C15A (3) or pET21a-SSU72-3319 (4) were run on SDS–PAGE gels. The position of protein Ssu72 is indicated by an asterisk. (B) Phosphatase activities present in samples (1 mg) of soluble crude extracts prepared from E.coli cells transformed with plasmid pET21a carrying either no insert (diamond), wild-type SSU72 gene (square), mutant ssu72-C15A (triangle) or mutant ssu72-3319 (cross). Samples were assayed for their ability to cleave pNPP (20 mM) after 20 min of incubation at 28°C. (C) Purification of His6-tagged Ssu72 protein. A sample of the purified protein was run on an SDS–PAGE gel. (D) Phosphatase activity of the purified His6-Ssu72 protein. Samples of purified Ssu72 (45 µg) were incubated as described in Figure 4A (diamond), in presence of 100 µg/ml BSA (square) or at 37°C (triangle). (E) Ortho-vanadate inhibition. Samples (45 µg) of purified His6-tagged Ssu72 protein were incubated with 20 mM pNPP during 20 min at 28°C, with an increasing concentration of ortho-vanadate.
Similar articles
- Functional interactions between the transcription and mRNA 3' end processing machineries mediated by Ssu72 and Sub1.
He X, Khan AU, Cheng H, Pappas DL Jr, Hampsey M, Moore CL. He X, et al. Genes Dev. 2003 Apr 15;17(8):1030-42. doi: 10.1101/gad.1075203. Genes Dev. 2003. PMID: 12704082 Free PMC article. - Ssu72 Is an RNA polymerase II CTD phosphatase.
Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Krishnamurthy S, et al. Mol Cell. 2004 May 7;14(3):387-94. doi: 10.1016/s1097-2765(04)00235-7. Mol Cell. 2004. PMID: 15125841 - Structurally conserved and functionally divergent yeast Ssu72 phosphatases.
Rodríguez-Torres AM, Lamas-Maceiras M, García-Díaz R, Freire-Picos MA. Rodríguez-Torres AM, et al. FEBS Lett. 2013 Aug 19;587(16):2617-22. doi: 10.1016/j.febslet.2013.06.044. Epub 2013 Jul 3. FEBS Lett. 2013. PMID: 23831060 - A structural perspective of CTD function.
Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. Meinhart A, et al. Genes Dev. 2005 Jun 15;19(12):1401-15. doi: 10.1101/gad.1318105. Genes Dev. 2005. PMID: 15964991 Review. - Diverse and conserved roles of the protein Ssu72 in eukaryotes: from yeast to higher organisms.
Liu C, Zhang W, Xing W. Liu C, et al. Curr Genet. 2021 Apr;67(2):195-206. doi: 10.1007/s00294-020-01132-5. Epub 2020 Nov 26. Curr Genet. 2021. PMID: 33244642 Review.
Cited by
- Mammalian Ssu72 phosphatase preferentially considers tissue-specific actively transcribed gene expression by regulating RNA Pol II transcription.
Kim HS, Jeon Y, Jang YO, Lee H, Shin Y, Lee CW. Kim HS, et al. Theranostics. 2022 Jan 1;12(1):186-206. doi: 10.7150/thno.62274. eCollection 2022. Theranostics. 2022. PMID: 34987641 Free PMC article. - Chemical Tools To Decipher Regulation of Phosphatases by Proline Isomerization on Eukaryotic RNA Polymerase II.
Mayfield JE, Fan S, Wei S, Zhang M, Li B, Ellington AD, Etzkorn FA, Zhang YJ. Mayfield JE, et al. ACS Chem Biol. 2015 Oct 16;10(10):2405-14. doi: 10.1021/acschembio.5b00296. Epub 2015 Sep 15. ACS Chem Biol. 2015. PMID: 26332362 Free PMC article. - Human snRNA genes use polyadenylation factors to promote efficient transcription termination.
O'Reilly D, Kuznetsova OV, Laitem C, Zaborowska J, Dienstbier M, Murphy S. O'Reilly D, et al. Nucleic Acids Res. 2014 Jan;42(1):264-75. doi: 10.1093/nar/gkt892. Epub 2013 Oct 4. Nucleic Acids Res. 2014. PMID: 24097444 Free PMC article. - The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3.
Cheng H, He X, Moore C. Cheng H, et al. Mol Cell Biol. 2004 Apr;24(7):2932-43. doi: 10.1128/MCB.24.7.2932-2943.2004. Mol Cell Biol. 2004. PMID: 15024081 Free PMC article. - The hsSsu72 phosphatase is a cohesin-binding protein that regulates the resolution of sister chromatid arm cohesion.
Kim HS, Baek KH, Ha GH, Lee JC, Kim YN, Lee J, Park HY, Lee NR, Lee H, Cho Y, Lee CW. Kim HS, et al. EMBO J. 2010 Oct 20;29(20):3544-57. doi: 10.1038/emboj.2010.217. Epub 2010 Sep 3. EMBO J. 2010. PMID: 20818333 Free PMC article.
References
- Cuff J.A., Clamp,M.E., Siddiqui,A.S., Finlay,M. and Barton,G.J. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics, 14, 892–893. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases