Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development - PubMed (original) (raw)
Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development
Yoshimi Tokuzawa et al. Mol Cell Biol. 2003 Apr.
Abstract
Embryonic stem (ES) cells are immortal and pluripotent cells derived from early mammalian embryos. Transcription factor Oct3/4 is essential for self-renewal of ES cells and early mouse development. However, only a few Oct3/4 target genes have been identified. In this study, we found that F-box-containing protein Fbx15 was expressed predominantly in mouse undifferentiated ES cells. Inactivation of Oct3/4 in ES cells led to rapid extinction of Fbx15 expression. Reporter gene analyses demonstrated that this ES cell-specific expression required an 18-bp enhancer element located approximately 500 nucleotides upstream from the transcription initiation site. The enhancer contained an octamer-like motif and an adjacent Sox-binding motif. Deletion or point mutation of either motif abolished the enhancer activity. The 18-bp fragment became active in NIH 3T3 cells when Oct3/4 and Sox2 were coexpressed. A gel mobility shift assay demonstrated cooperative binding of Oct3/4 and Sox2 to the enhancer sequence. In mice having a beta-galactosidase gene knocked into the Fbx15 locus, 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside staining was detected in ES cells, early embryos (two-cell to blastocyst stages), and testis tissue. Despite such specific expression of Fbx15, homozygous mutant mice showed no gross developmental defects and were fertile. Fbx15-null ES cells were normal in morphology, proliferation, and differentiation. These data demonstrate that Fbx15 is a novel target of Oct3/4 but is dispensable for ES cell self-renewal, development, and fertility.
Figures
FIG. 1.
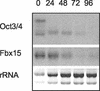
(A) RT-PCR analysis showing the expression profiles of mouse Oct3/4, Fbx15, and NAT1. Lanes: a, undifferentiated ES cells; b, differentiated ES cells; c, brain; d, heart; e, kidney; f, testis; g, spleen; h, muscle; i, lung; j, stomach; k, ovary; l, thymus; m, liver; n, skin. (B) Western blot analysis showing Fbx15 expression in the RF8, J1, CGR8, and MG1.19 ES cell lines. Lysates were collected from ES cells maintained undifferentiated (lanes 0) or treated with retinoic acid for 5 days (lanes 5). Western blotting was performed with anti-Fbx15 serum or anti-CDK4 antibody.
FIG. 2.
Expression of Fbx15 in ZHBTc4 ES cells. Cells were treated with tetracycline, and the expression of Oct3/4 and Fbx15 was determined at the indicated time points by Northern blot analysis. Ethidium bromide staining of rRNAs was used as a loading control.
FIG. 3.
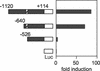
Reporter gene analyses to define the Fbx15 enhancer. DNA fragments starting at positions −1120, −640, and −526 and ending at position +114 of the Fbx15 gene were placed in front of the firefly luciferase gene. These reporter constructs were introduced into undifferentiated ES cells. Shown is fold induction of normalized luciferase activity compared to that of the promoterless construct (pGV-BM2). Diagonally striped boxes represent the octamer-like and Sox-binding motifs.
FIG. 4.
Enhancer analyses with the minimal thymidine kinase (TK) promoter. DNA fragments of various sizes were isolated from the Fbx15 gene and placed in front of the firefly luciferase gene driven by the minimal thymidine kinase promoter. These reporter constructs were introduced into undifferentiated ES cells (black column), differentiated ES cells (gray), and NIH 3T3 cells (white). Shown is fold induction of normalized luciferase activity compared to that of the enhancerless construct (pTA-Luc). Diagonally striped boxes represent the octamer-like and Sox-binding motifs.
FIG. 5.
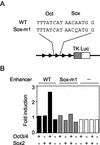
Effects of point mutations in the octamer-like sequence or the Sox recognition motif. Reporter plasmids containing the Fbx15 enhancer sequence (WT) or mutated sequences (Oct-m, Sox-m1, and Sox-m2) were analyzed as described in the legend to Fig. 4.
FIG. 6.
Activation of the Fbx15 enhancer by Oct3/4 and Sox2 in NIH 3T3 cells. Reporter plasmids containing five copies of the Fbx15 enhancer sequence (WT) or a mutated sequence (Sox-m1) were introduced into NIH 3T3 cells with or without Oct3/4 and/or Sox2 expression vectors. Luciferase activities were analyzed as described in the legend to Fig. 4.
FIG. 7.
Gel mobility shift assay. A 32P-labeled oligonucleotide (WT; ccagatgtgcTTTATCATAACAATGgaattcctaggggct) corresponding to the Fbx15 enhancer was incubated with F9 EC cell extract or Cos7 cell extract expressing Oct3/4, Sox2, or both. Oligonucleotides with mutations (underlined) in the octamer-like sequence [O (−); ccagatgtgcT
CCC
TCATAACAATGgaattcctaggggct] and the Sox-binding site [S (−); ccagatgtgcTTTATCATAAC
C
ATGgaattcctaggggct] were also tested. As a control, an FGF-4 enhancer oligonucleotide (tttaagtatcccATTAGCATccaAACAAAGagttttcta) was incubated with Cos7 cell extracts expressing Oct3/4, Sox2, or both. Shown on the left are the positions of bands corresponding to each transcription factor(s). The nature of the faint band indicated by the asterisk is not known. cold WT, unlabeled oligonucleotide corresponding to the Fbx15 enhancer.
FIG. 8.
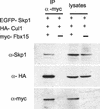
Western blot analyses showing association of Fbx15 with Skp1 and Cul1. MG1.19 cells were transfected with myc-Fbx15, HA-Cul1, and EGFP-Skp1 expression vectors. Cell lysates and anti-myc antibody immunoprecipitates (IP) were separated by SDS-PAGE and immunoblotted with anti-Skp1 (top), anti-HA (middle), and anti-myc (bottom) antibodies. In a negative-control experiment, myc-Fbx15 was omitted from the transfection reaction mixture.
FIG. 9.
Targeted disruption of the mouse Fbx15 gene. (A) Structures of the Fbx15 genomic locus, a targeting vector, and the targeted locus generated by homologous recombination. The targeting vector contains the β-geo cassette in place of exons 3 to 7. The length of the diagnostic _Hin_dIII (H) restriction fragments and the locations of the 5′ and 3′ probes for Southern blot analyses are shown. Arrows indicate the primers for PCR analysis. The diagrams are not drawn to scale. (B) Southern blot analysis. Specific hybridization with the 5′ probe produces a 10-kb band from the wild-type locus and an 8-kb band from the targeted locus. Hybridization with the 3′ probe produces a 9-kb band from the wild-type locus and a 10-kb band from the targeted locus. +/+ and ± represent genotypes of Fbx15+/+ and Fbx15+/− cells, respectively. (C) PCR analysis. PCR with the three primers shown in panel A produces a 280-bp band form the wild-type locus and a 725-bp band from the targeted locus. (D) Northern blot analysis. Total RNA isolated from ES cells of each genotype was separated, blotted, and hybridized to cDNA probes of Fbx15 and Oct3/4. (E) Western blot analyses. Lysates isolated from ES cells of each genotype were separated by SDS-PAGE and immunoblotted with anti-Fbx15 serum or anti-CDK4 antibody.
FIG. 10.
Fbx15 expression represented by β-gal activity in mutant ES cells and mice. (A) Wild-type and _Fbx15_−/− ES cells were stained with X-Gal. Cells were either maintained undifferentiated on feeder cells or induced to differentiate with retinoic acid. (B) Whole-mount X-Gal staining of testes from wild-type and Fbx15+/− mice. (C) Whole-mount X-Gal staining of eggs from Fbx15+/+ and Fbx15+/− female mice mated with wild-type males.
Similar articles
- Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells.
Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Masui S, et al. Nat Cell Biol. 2007 Jun;9(6):625-35. doi: 10.1038/ncb1589. Epub 2007 May 21. Nat Cell Biol. 2007. PMID: 17515932 - Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells.
Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Maruyama M, et al. J Biol Chem. 2005 Jul 1;280(26):24371-9. doi: 10.1074/jbc.M501423200. Epub 2005 Apr 29. J Biol Chem. 2005. PMID: 15863505 - Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells.
Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Okumura-Nakanishi S, et al. J Biol Chem. 2005 Feb 18;280(7):5307-17. doi: 10.1074/jbc.M410015200. Epub 2004 Nov 22. J Biol Chem. 2005. PMID: 15557334 - Animal embryonic stem (ES) cells: self-renewal, pluripotency, transgenesis and nuclear transfer.
Saito S, Liu B, Yokoyama K. Saito S, et al. Hum Cell. 2004 Sep;17(3):107-15. doi: 10.1111/j.1749-0774.2004.tb00026.x. Hum Cell. 2004. PMID: 15859155 Review. - Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation.
Kondoh H, Uchikawa M, Kamachi Y. Kondoh H, et al. Int J Dev Biol. 2004;48(8-9):819-27. doi: 10.1387/ijdb.041868hk. Int J Dev Biol. 2004. PMID: 15558474 Review.
Cited by
- Roles of F-box proteins in cancer.
Wang Z, Liu P, Inuzuka H, Wei W. Wang Z, et al. Nat Rev Cancer. 2014 Apr;14(4):233-47. doi: 10.1038/nrc3700. Nat Rev Cancer. 2014. PMID: 24658274 Free PMC article. Review. - ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency.
Iwabuchi KA, Yamakawa T, Sato Y, Ichisaka T, Takahashi K, Okita K, Yamanaka S. Iwabuchi KA, et al. PLoS One. 2011;6(5):e20461. doi: 10.1371/journal.pone.0020461. Epub 2011 May 26. PLoS One. 2011. PMID: 21637830 Free PMC article. - A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells.
Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. Fong YW, et al. Cell. 2011 Sep 30;147(1):120-31. doi: 10.1016/j.cell.2011.08.038. Cell. 2011. PMID: 21962512 Free PMC article. - Transcription factor heterogeneity and epiblast pluripotency.
Osorno R, Chambers I. Osorno R, et al. Philos Trans R Soc Lond B Biol Sci. 2011 Aug 12;366(1575):2230-7. doi: 10.1098/rstb.2011.0043. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 21727128 Free PMC article. - Parthenogenesis as an approach to pluripotency: advantages and limitations involved.
Brevini TA, Pennarossa G, Antonini S, Gandolfi F. Brevini TA, et al. Stem Cell Rev. 2008 Sep;4(3):127-35. doi: 10.1007/s12015-008-9027-z. Epub 2008 Jun 12. Stem Cell Rev. 2008. PMID: 18548354 Review.
References
- Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. - PubMed
- Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. - PMC - PubMed
- Bodine, S. C., E. Latres, S. Baumhueter, V. K. Lai, L. Nunez, B. A. Clarke, W. T. Poueymirou, F. J. Panaro, E. Na, K. Dharmarajan, Z. Q. Pan, D. M. Valenzuela, T. M. DeChiara, T. N. Stitt, G. D. Yancopoulos, and D. J. Glass. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294:1704-1708. - PubMed
- Damjanov, I. 1978. Teratoma and teratocarcinoma in experimental animals. Natl. Cancer Inst. Monogr. 49:305-306. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous