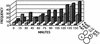Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion - PubMed (original) (raw)
Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion
Shaune Edwards et al. Mol Cell Biol. 2003 Apr.
Abstract
The large subunit of Saccharomyces cerevisiae DNA polymerase epsilon, Pol2, comprises two essential functions. The N terminus has essential DNA polymerase activity. The C terminus is also essential, but its function is unknown. We report here that the C-terminal domain of Pol2 interacts with polymerase sigma (Pol sigma), a recently identified, essential nuclear nucleotidyl transferase encoded by two redundant genes, TRF4 and TRF5. This interaction is functional, since Pol sigma stimulates the polymerase activity of the Pol epsilon holoenzyme significantly. Since Trf4 is required for sister chromatid cohesion as well as for completion of S phase and repair, the interaction suggested that Pol epsilon, like Pol sigma, might form a link between the replication apparatus and sister chromatid cohesion and/or repair machinery. We present evidence that pol2 mutants are defective in sister chromatid cohesion. In addition, Pol2 interacts with SMC1, a subunit of the cohesin complex, and with ECO1/CTF7, required for establishing sister chromatid cohesion; and pol2 mutations act synergistically with smc1 and scc1. We also show that trf5 Delta mutants, like trf4 Delta mutants, are defective in DNA repair and sister chromatid cohesion.
Figures
FIG. 1.
(A) Schematic diagram of the POL2 gene describing conserved exonuclease domains and polymerase domains. The C-terminal portion of Pol2 used for the two-hybrid library screen includes aa 1265 to 2222. Ten amino acid deletions in the zinc finger region of Pol2 were used to map the interaction of Trf5; the zinc finger deletion region is shown enlarged. (B) Schematic diagram of the TRF5 gene. The conserved nucleotidyl transferase domain occurs between aa 178 and 241. The principle conserved motif is from aa 217 to 235. Aspartates 233 and 235 in Trf5 correspond to aspartates 236 and 238 in the DXD motif in Trf4, and since mutations in these residues reduce the DNA polymerase activity of Trf4, we have labeled this the putative active site (6, 70). (C) The 78-aa POL2 interaction region of TRF5 homologous to TRF genes in yeast, human, and fly. The alignment was made using BLAST data from the SGD and GenBank. SC, S. cerevisiae; SP_, Schizosaccharomyces pombe_; DM, Drosophila melanogaster; HS, Homo sapiens. The corresponding amino acid numbers and accession numbers are SC-Trf5_1050861 (aa 371 to 448); SC-Trf4_950226 (aa 374 to 451); SP-C12G12.13C_2130260 (aa 1006 to 1081); DM-CG11265_22831959 (aa 461 to 538); HS-Trf4_5565687 (aa 105 to 182); HS-Lak-1_5139669 (aa 181 to 258); A. gambiae_21288943 (aa 460 to 536). Both identities and similarities are highlighted. The asterisks identify mutations in trf4 that cause lethality (trf4-378; trf4-425; trf4-444).
FIG. 2.
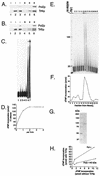
(A) Pol2 coimmunoprecipitates with Trf4. Coexpression of FLAG-Pol2 and His-Trf4 and immunoprecipitation protocols are described in Materials and Methods. Western blots of crude extract (labeled “I” for input protein) from insect cells expressing various combinations of Trf proteins and FLAG-Pol2 were probed with antibody against Trf4 or anti-FLAG Pol2 as indicated. Lane 1, no recombinant protein; lane 2, His-Trf4; lane 3, His-Trf4 plus FLAG-Pol2. These extracts were incubated with anti-FLAG beads. After washing of the beads, proteins that bound from extracts (labeled “B” for bound protein) were eluted by boiling. The proteins were analyzed on Western blots probed with antibody against Trf4 or anti-FLAG Pol2 as indicated on the right. Lane 4, no recombinant protein; lane 5, His-Trf4; lane 6, His-Trf4 plus FLAG-Pol2. (B) Pol2 coimmunoprecipitates with Trf5. Coexpression of FLAG-Pol2 and Trf5-His is described in Materials and Methods. Western blots of crude extract from insect cells are represented in the same order as those in Fig. 2A. (C) Recombinant Trf4 prepared in E. coli stimulates Pol ɛ holoenzyme. Trf4 was purified exactly as described previously (70). The oligo(dT)12-18 primer extension assay is described in Materials and Methods. Reaction mixtures contained 680 ng of Trf4, the amount required to observe Trf4 DNA polymerase activity (lanes 2 and 3), and/or 0.15 U of Pol ɛ, as indicated, are shown. The high level of Trf4 is saturating for stimulatory activity (see panel D). Lane 1, no protein; lane 2, Trf4-His with 0.1 mM dTTP; lane 3, Trf4-His with 1 mM dTTP; lane 4, Pol ɛ with 0.1 mM dTTP; lane 5, Pol ɛ with 1 mM dTTP; lane 6, Pol ɛ plus Trf4-His with 0.1 mM dTTP; and lane 7, Pol ɛ plus Trf4-His with 1 mM dTTP. (D) Titration of stimulatory activity of scTrf4 made in bacteria. The indicated amounts of scTrf4 were assayed for stimulation of [3H]dTMP incorporation by 0.15 U of Pol ɛ on an oligo(dT)-poly(dA) substrate as described in Materials and Methods. (E) scTrf4-His expressed in insect cell cochromatographs with Pol ɛ-stimulatory activity. scTrf4-His was expressed in insect cells. Silver staining of Trf4-His after purification through Ni2+-nitrilotriacetic acid and Mono Q columns, as described in Materials and Methods, and gel electrophoresis is shown at the top. Numbers refer to MonoQ fraction numbers. The same fractions are assayed for stimulation of primer extension by pol ɛ. Each fraction from the Mono Q column was dialyzed, and 2 μl of each fraction was used in a 20-μl reaction. Fraction 13 contained 17 ng of Trf4 protein (13 nM); but 2.7 nM Trf4 gave equivalent stimulation (not shown). The first lane shows no primer extension, the second lane shows activity of 0.15 U of Pol ɛ (0.5 nM) alone, and the subsequent lanes are the Mono Q fractions of the Trf4 purification assayed with 0.15 U of Pol ɛ (0.5 nm). The fraction numbers are identified above. (F) scTrf4-His and Pol ɛ-stimulatory activity copurify. [3H]dTMP incorporation assay: the same fractions from the Mono Q column were assayed for [3H]dTMP incorporation on an oligo(dT)-poly(dA) substrate as described in Materials and Methods in the presence of 0.15 U of Pol ɛ (0.5 nm). (G) Trf4 from the MonoQ column is highly purified. Coomassie-stained gel of Trf4 from fraction 14 of the MonoQ column. (H) scTrf4-His does not efficiently stimulate Pol2-140 lacking the C-terminal 1,000 amino acids. Three levels (0.075, 0.15, or 0.3 U) of either Pol ɛ (solid dots) or truncated Pol2 protein (open dots) were assayed with saturating amounts (40 ng) of Trf4-His purified from insect cells. Similar results were obtained with scTrf4 prepared in E. coli.
FIG. 3.
Genetic interaction of POL2 and TRF4. POL2 and DPB2 suppress the cold sensitivity in a trf4Δ mutant. Plasmids PDLT4, pSEY18-II, pSEY18-DPB2, and pSEY18 (GAL1 and GAL10 expression vectors with inserts of TRF4, POL2, DPB2, or empty [no insert], respectively) were transformed into strains 6265 (BY4741-trf4Δ) and the wild type (BY4741) and selected for growth on synthetic medium with dextrose-Ura. The resultant colonies were purified, grown to log phase in liquid SRaff-URA, and serially diluted onto plates containing 2% galactose. Duplicate plates were incubated at 16°C for a week.
FIG. 4.
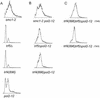
Effects of combining mutations in trf, pol2, and smc genes. (A) Flow cytometry profile of asynchronous cultures of exponentially growing pol2-12, trf4(896), trf5Δ, and smc1-2 single mutants_._ (B) Double mutants pol2-12 trf4(896) and _pol2-12 trf5_Δ. (C) Two pol2-12 trf4(896) trf5 triple mutants (TM6 and TM3). All strains were generated by the crosses described in Materials and Methods. Cells were grown to log phase (approximately 2 × 107 cells/ml) at 30°C; however, strains exhibiting an extended S phase were less dense.
FIG. 5.
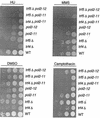
Sensitivity of pol2-11, pol2-12, trf4, and trf5 mutants and various combination mutants to DNA synthesis inhibitors and DNA damaging reagents. Strains were grown to log phase and serially diluted at 104, 103, 102, and 10 cells per row on YPD with dimethyl sulfoxide (DMSO) (control plates, DMSO was used to dilute camptothecin; see Materials and Methods), 10-μg/ml camptothecin, 125 mM HU, and 0.03% (vol/vol) MMS. Strains are isogenic with the following designations: WT, BY4741; _trf4_Δ, 6265; _trf5_Δ, 1145; pol2-11, BY-pol2-11; pol2-12, BY-pol2-12; _trf4_Δ pol2-11, BY-trf4_Δ_pol2-11; _trf5_Δ pol2-11, BY-trf5_Δ_pol2-11; and _trf5_Δ pol2-12, BY-trf5_Δ_pol2-12 (see Table 1).
FIG. 6.
Defective sister chromatid cohesion observed in both a pol2-12 mutant and a trf5Δ mutant. (A) Flow-cytometric analysis of nocodazole-arrested pol2-12 mutant and an isogenic wild-type strain carrying the GFP signal. (B) Typical cells with attached sisters (one GFP dot per cell body) or separated sisters (two separated dots per cell body). (C) Defective sister chromatid separation was measured as total GFP dots, relative to double GFP dots (dd), for 500 to 700 cells in two separate experiments for each strain. Both pol2-12 and trf5_Δ are defective. Replacing POL2 alleviates the defect. Isogenic controls for 12OG2(pADH-POL2) were wild type, SOG3, and 12OG2 (a pol2-12 mutant) (Table 1). These gave similar values of approximately 20% dd, similar to values observed with isogenic strains AFS479 and JCY122 shown here. Strains are designated as follows: AFS479 wild type (WT), JCY122 (pol2-12), JCYT59 (trf5Δ),_ 12OG2 [pol2-12(pADH-POL2)], JCYPD18 [cdc2-1 (Pol δ)], and JCYDNA2.
FIG. 7.
Sister chromatid separation in a pol2 mutant begins earlier in the cell cycle than in the wild type. Wild-type cells (AFS479) and pol2-12 cells (JCY122) were grown to log phase at 30°C, induced for GFP, and then arrested with α-factor for 3 h. Cells were then released in YPD at 30°C (A and B) or 34°C (C). Samples were taken every 15 min and analyzed for bud emergence by differential interference contrast (represented as budding index), timing of sister chromatid separation by GFP assay, spindle elongation by indirect immunofluorescence using an antitubulin antibody, DNA content by flow cytometry, and nuclear morphology by DAPI staining.
FIG. 8.
The frequency of separated sister chromatids occurring in a single nucleus increases for the pol2 mutant. Cells that displayed two GFP dots in Fig. 6 were again scored for the percentage of these that displayed two signals in one nucleus.
Similar articles
- Pol kappa: A DNA polymerase required for sister chromatid cohesion.
Wang Z, Castaño IB, De Las Peñas A, Adams C, Christman MF. Wang Z, et al. Science. 2000 Aug 4;289(5480):774-9. doi: 10.1126/science.289.5480.774. Science. 2000. PMID: 10926539 - Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion.
Skibbens RV. Skibbens RV. Genetics. 2004 Jan;166(1):33-42. doi: 10.1534/genetics.166.1.33. Genetics. 2004. PMID: 15020404 Free PMC article. - Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol sigma DNA polymerase.
Wang Z, Castaño IB, Adams C, Vu C, Fitzhugh D, Christman MF. Wang Z, et al. Genetics. 2002 Feb;160(2):381-91. doi: 10.1093/genetics/160.2.381. Genetics. 2002. PMID: 11861546 Free PMC article. - Evidence that replication fork components catalyze establishment of cohesion between sister chromatids.
Carson DR, Christman MF. Carson DR, et al. Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8270-5. doi: 10.1073/pnas.131022798. Proc Natl Acad Sci U S A. 2001. PMID: 11459963 Free PMC article. Review. - Replication-related activities establish cohesion between sister chromatids.
Wang Z, Christman MF. Wang Z, et al. Cell Biochem Biophys. 2001;35(3):289-301. doi: 10.1385/CBB:35:3:289. Cell Biochem Biophys. 2001. PMID: 11894848 Review.
Cited by
- R-loop mediated transcription-associated recombination in trf4Δ mutants reveals new links between RNA surveillance and genome integrity.
Gavaldá S, Gallardo M, Luna R, Aguilera A. Gavaldá S, et al. PLoS One. 2013 Jun 7;8(6):e65541. doi: 10.1371/journal.pone.0065541. Print 2013. PLoS One. 2013. PMID: 23762389 Free PMC article. - Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity.
Haracska L, Johnson RE, Prakash L, Prakash S. Haracska L, et al. Mol Cell Biol. 2005 Nov;25(22):10183-9. doi: 10.1128/MCB.25.22.10183-10189.2005. Mol Cell Biol. 2005. PMID: 16260630 Free PMC article. - A network of multi-tasking proteins at the DNA replication fork preserves genome stability.
Budd ME, Tong AH, Polaczek P, Peng X, Boone C, Campbell JL. Budd ME, et al. PLoS Genet. 2005 Dec;1(6):e61. doi: 10.1371/journal.pgen.0010061. Epub 2005 Dec 2. PLoS Genet. 2005. PMID: 16327883 Free PMC article. - Understanding and predicting synthetic lethal genetic interactions in Saccharomyces cerevisiae using domain genetic interactions.
Li B, Cao W, Zhou J, Luo F. Li B, et al. BMC Syst Biol. 2011 May 17;5:73. doi: 10.1186/1752-0509-5-73. BMC Syst Biol. 2011. PMID: 21586150 Free PMC article. - Mutations in the Non-Catalytic Subunit Dpb2 of DNA Polymerase Epsilon Affect the Nrm1 Branch of the DNA Replication Checkpoint.
Dmowski M, Rudzka J, Campbell JL, Jonczyk P, Fijałkowska IJ. Dmowski M, et al. PLoS Genet. 2017 Jan 20;13(1):e1006572. doi: 10.1371/journal.pgen.1006572. eCollection 2017 Jan. PLoS Genet. 2017. PMID: 28107343 Free PMC article.
References
- Aparicio, O. M., D. M. Weinstein, and S. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases