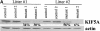Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A - PubMed (original) (raw)
Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A
Chun-Hong Xia et al. J Cell Biol. 2003.
Abstract
To test the hypothesis that fast anterograde molecular motor proteins power the slow axonal transport of neurofilaments (NFs), we used homologous recombination to generate mice lacking the neuronal-specific conventional kinesin heavy chain, KIF5A. Because null KIF5A mutants die immediately after birth, a synapsin-promoted Cre-recombinase transgene was used to direct inactivation of KIF5A in neurons postnatally. Three fourths of such mutant mice exhibited seizures and death at around 3 wk of age; the remaining animals survived to 3 mo or longer. In young mutant animals, fast axonal transport appeared to be intact, but NF-H, as well as NF-M and NF-L, accumulated in the cell bodies of peripheral sensory neurons accompanied by a reduction in sensory axon caliber. Older animals also developed age-dependent sensory neuron degeneration, an accumulation of NF subunits in cell bodies and a reduction in axons, loss of large caliber axons, and hind limb paralysis. These data support the hypothesis that a conventional kinesin plays a role in the microtubule-dependent slow axonal transport of at least one cargo, the NF proteins.
Figures
Figure 1.
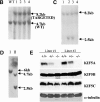
Targeted disruption of the mouse KIF5A gene. (A) KIF5A gene targeting strategy. In the targeting vector, the pGK-neo and HSV-tk selection cassette was flanked by two loxP sites (loxP1 and loxP2), and the two exons to be deleted were flanked by loxP2 and loxP3. Two steps of transfection were performed to generate type I deletion (null mutant) and type II deletion ES cells. The first step was done by transfecting linearized targeting vector into ES cells followed by G418 selection; the second step was performed by transfecting a Cre plasmid into recombinant ES cells isolated from the first step, followed by Gancyclovir selection. (B and C) Southern blot analyses of HindIII-digested G418-resistant ES clones after transfecting targeting vector. (B) A 5′ external probe was used to identify the recombinant clones. A 4.7-kb wild-type band was detected, and an additional 8.2 kb (with the addition of a 3.5-kb selection cassette) was also detected in the recombinant clones. (C) The presence of three loxP sites was confirmed by a loxP probe. The correct recombinant clones should have all three loxP sites, both 8.2-kb and 2.5-kb bands should be detected by the probe (clones 2 and 3), whereas clones 1 and 4 only had the first two loxP sites. (D) Southern blot analysis of HindIII-digested Gancyclovir-resistant clones after Cre transfection. With the loxP probe, only a 6-kb band was detected in type I deletion (KIF5A_null_) ES cells, whereas 4.7-kb and 2.5-kb bands were detected in type II (KIF5A_flox_) ES cells. (E) No KIF5A protein was detected in KIF5A null mutant mice by Western blot analysis. Mouse brain homogenates were made from two litters of mice, and 100 μg of protein was loaded in each lane. Isoform-specific antibodies were used to probe KIF5A, KIF5B, and KIF5C; α-tubulin was used as a loading control.
Figure 1.
Targeted disruption of the mouse KIF5A gene. (A) KIF5A gene targeting strategy. In the targeting vector, the pGK-neo and HSV-tk selection cassette was flanked by two loxP sites (loxP1 and loxP2), and the two exons to be deleted were flanked by loxP2 and loxP3. Two steps of transfection were performed to generate type I deletion (null mutant) and type II deletion ES cells. The first step was done by transfecting linearized targeting vector into ES cells followed by G418 selection; the second step was performed by transfecting a Cre plasmid into recombinant ES cells isolated from the first step, followed by Gancyclovir selection. (B and C) Southern blot analyses of HindIII-digested G418-resistant ES clones after transfecting targeting vector. (B) A 5′ external probe was used to identify the recombinant clones. A 4.7-kb wild-type band was detected, and an additional 8.2 kb (with the addition of a 3.5-kb selection cassette) was also detected in the recombinant clones. (C) The presence of three loxP sites was confirmed by a loxP probe. The correct recombinant clones should have all three loxP sites, both 8.2-kb and 2.5-kb bands should be detected by the probe (clones 2 and 3), whereas clones 1 and 4 only had the first two loxP sites. (D) Southern blot analysis of HindIII-digested Gancyclovir-resistant clones after Cre transfection. With the loxP probe, only a 6-kb band was detected in type I deletion (KIF5A_null_) ES cells, whereas 4.7-kb and 2.5-kb bands were detected in type II (KIF5A_flox_) ES cells. (E) No KIF5A protein was detected in KIF5A null mutant mice by Western blot analysis. Mouse brain homogenates were made from two litters of mice, and 100 μg of protein was loaded in each lane. Isoform-specific antibodies were used to probe KIF5A, KIF5B, and KIF5C; α-tubulin was used as a loading control.
Figure 2.
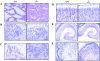
Histology of KIF5A null mutant mice. (A) Lung histology of KIF5A null mutant. 7-μm lung paraffin sections from KIF5A null (KIF5A_null_/KIF5A_null_) and control (KIF5A_WT_/KIF5A_WT_) littermates were stained with hematoxylin and eosin. Note that the mutant lung was not well expanded. Bar, 50 μm. (B–F) Histology of KIF5A null mutant nervous tissues. Paraffin sections from spinal cord (B and C), cortex (D), hippocampus (E), and cerebellum (F) were stained with cresyl violet. Note that no obvious differences were observed between KIF5A null mutant and control littermates except that the cell bodies of the motor neurons were larger in the mutant spinal cord. Bars: (B, D, and F) 50 μm; (C) 20 μm; (E) 100 μm.
Figure 3.
Gross analysis of KIF5A conditional mutant mice (KIF5A nul l/KIF5A_flox_; Cre_synapsin_). (A) Western blot analysis to measure KIF5A protein levels in the brains of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mice. Equal amounts of brain homogenate from 3-wk-old mutant and control (KIF5A_flox_/KIF5A_WT_) littermates were loaded; actin was used as a loading control. Each marked number represents the ratio between the mutant band and control band after normalizing with the actin band. (B and C) KIF5A excision by Cre_synapsin_ transgene in DRG (B) and spinal cord motor neurons (C) of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant. Tissue sections from ∼3-wk-old mutant and control littermates were stained with KIF5A-specific antibody. Spinal cord sections were also double stained with an anti-BIP antibody to visualize the motor neuron cell bodies. Note the decreased or lack of KIF5A staining in some neurons (arrowheads). Bar, 100 μm. (D) A comparison of body weight (mean ± SD) among 3-wk-old littermates with different genotypes. Note the obvious low body weight in the KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant group, n = 4 for each group. *, P < 0.01. (E) Most KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice died around 3 wk of age. Postnatal survival curve of a group of mice (142 total, 113 control and 29 mutant) is shown here. The rate of survival of the different genotypes was plotted against age. (F) Abnormal hind limb posture in an older KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mouse. Two 7.5-mo-old littermates (control and KIF5A_null_/KIF5A_flox_) are shown.
Figure 3.
Gross analysis of KIF5A conditional mutant mice (KIF5A nul l/KIF5A_flox_; Cre_synapsin_). (A) Western blot analysis to measure KIF5A protein levels in the brains of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mice. Equal amounts of brain homogenate from 3-wk-old mutant and control (KIF5A_flox_/KIF5A_WT_) littermates were loaded; actin was used as a loading control. Each marked number represents the ratio between the mutant band and control band after normalizing with the actin band. (B and C) KIF5A excision by Cre_synapsin_ transgene in DRG (B) and spinal cord motor neurons (C) of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant. Tissue sections from ∼3-wk-old mutant and control littermates were stained with KIF5A-specific antibody. Spinal cord sections were also double stained with an anti-BIP antibody to visualize the motor neuron cell bodies. Note the decreased or lack of KIF5A staining in some neurons (arrowheads). Bar, 100 μm. (D) A comparison of body weight (mean ± SD) among 3-wk-old littermates with different genotypes. Note the obvious low body weight in the KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant group, n = 4 for each group. *, P < 0.01. (E) Most KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice died around 3 wk of age. Postnatal survival curve of a group of mice (142 total, 113 control and 29 mutant) is shown here. The rate of survival of the different genotypes was plotted against age. (F) Abnormal hind limb posture in an older KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mouse. Two 7.5-mo-old littermates (control and KIF5A_null_/KIF5A_flox_) are shown.
Figure 3.
Gross analysis of KIF5A conditional mutant mice (KIF5A nul l/KIF5A_flox_; Cre_synapsin_). (A) Western blot analysis to measure KIF5A protein levels in the brains of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mice. Equal amounts of brain homogenate from 3-wk-old mutant and control (KIF5A_flox_/KIF5A_WT_) littermates were loaded; actin was used as a loading control. Each marked number represents the ratio between the mutant band and control band after normalizing with the actin band. (B and C) KIF5A excision by Cre_synapsin_ transgene in DRG (B) and spinal cord motor neurons (C) of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant. Tissue sections from ∼3-wk-old mutant and control littermates were stained with KIF5A-specific antibody. Spinal cord sections were also double stained with an anti-BIP antibody to visualize the motor neuron cell bodies. Note the decreased or lack of KIF5A staining in some neurons (arrowheads). Bar, 100 μm. (D) A comparison of body weight (mean ± SD) among 3-wk-old littermates with different genotypes. Note the obvious low body weight in the KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant group, n = 4 for each group. *, P < 0.01. (E) Most KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice died around 3 wk of age. Postnatal survival curve of a group of mice (142 total, 113 control and 29 mutant) is shown here. The rate of survival of the different genotypes was plotted against age. (F) Abnormal hind limb posture in an older KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mouse. Two 7.5-mo-old littermates (control and KIF5A_null_/KIF5A_flox_) are shown.
Figure 3.
Gross analysis of KIF5A conditional mutant mice (KIF5A nul l/KIF5A_flox_; Cre_synapsin_). (A) Western blot analysis to measure KIF5A protein levels in the brains of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mice. Equal amounts of brain homogenate from 3-wk-old mutant and control (KIF5A_flox_/KIF5A_WT_) littermates were loaded; actin was used as a loading control. Each marked number represents the ratio between the mutant band and control band after normalizing with the actin band. (B and C) KIF5A excision by Cre_synapsin_ transgene in DRG (B) and spinal cord motor neurons (C) of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant. Tissue sections from ∼3-wk-old mutant and control littermates were stained with KIF5A-specific antibody. Spinal cord sections were also double stained with an anti-BIP antibody to visualize the motor neuron cell bodies. Note the decreased or lack of KIF5A staining in some neurons (arrowheads). Bar, 100 μm. (D) A comparison of body weight (mean ± SD) among 3-wk-old littermates with different genotypes. Note the obvious low body weight in the KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant group, n = 4 for each group. *, P < 0.01. (E) Most KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice died around 3 wk of age. Postnatal survival curve of a group of mice (142 total, 113 control and 29 mutant) is shown here. The rate of survival of the different genotypes was plotted against age. (F) Abnormal hind limb posture in an older KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mouse. Two 7.5-mo-old littermates (control and KIF5A_null_/KIF5A_flox_) are shown.
Figure 3.
Gross analysis of KIF5A conditional mutant mice (KIF5A nul l/KIF5A_flox_; Cre_synapsin_). (A) Western blot analysis to measure KIF5A protein levels in the brains of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mice. Equal amounts of brain homogenate from 3-wk-old mutant and control (KIF5A_flox_/KIF5A_WT_) littermates were loaded; actin was used as a loading control. Each marked number represents the ratio between the mutant band and control band after normalizing with the actin band. (B and C) KIF5A excision by Cre_synapsin_ transgene in DRG (B) and spinal cord motor neurons (C) of KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant. Tissue sections from ∼3-wk-old mutant and control littermates were stained with KIF5A-specific antibody. Spinal cord sections were also double stained with an anti-BIP antibody to visualize the motor neuron cell bodies. Note the decreased or lack of KIF5A staining in some neurons (arrowheads). Bar, 100 μm. (D) A comparison of body weight (mean ± SD) among 3-wk-old littermates with different genotypes. Note the obvious low body weight in the KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant group, n = 4 for each group. *, P < 0.01. (E) Most KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice died around 3 wk of age. Postnatal survival curve of a group of mice (142 total, 113 control and 29 mutant) is shown here. The rate of survival of the different genotypes was plotted against age. (F) Abnormal hind limb posture in an older KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mouse. Two 7.5-mo-old littermates (control and KIF5A_null_/KIF5A_flox_) are shown.
Figure 4.
Sensory axon degeneration in KIF5A null /KIF5A_flox_; Cre_synapsin_ mutant mice. (A) Ventral roots of 3-wk-old and 5.5-mo-old mutant mice. Note the lack of degeneration. Bar, 50 μm. (B) Dorsal roots of 3-wk-old and 5.5-mo-old mutant mice. Note the lack of degeneration in 3-wk sensory axons, but in the 5.5-mo-old mutant, there are large numbers of degenerating axons (arrowheads) and a striking decrease of large caliber axons. Bar, 20 μm.
Figure 5.
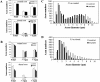
Loss of large caliber axons in the KIF5A null /KIF5A_flox_; Cre_synapsin_ mutant mice. (A) Counts of total, small (<3 μm diameter), and large (>3 μm diameter) myelinated axons in L5 ventral and dorsal roots from 3-wk-old control (n = 4) and mutant (n = 6) mice. No significant difference is observed between the number of mutant and control axons in the ventral root. There is a significant loss of dorsal root axons. *, P < 0.05 (0.045); **, P < 0.01 (0.002); _t_ test. (B) Axon numbers of total, small (<4 μm), and large (>4 μm) myelinated axons in L5 ventral and dorsal roots from 5.5-mo-old control (1 and 2, control) and mutant (3 and 4, mutant) mice. There is a profound loss of dorsal root axons, especially large caliber axons. The loss of ventral root axons is not as profound as observed in the dorsal root. (C and D) Axonal diameters of L5 ventral root (C) and L5 dorsal root (D) from 5.5-mo-old mutant and control mice. Averaged distribution of axon diameters from the entire roots of two mutant and two control mice is shown. Note that in the ventral root (C), the peak of the distribution of the diameter of large caliber axons shifts from 7–7.5 μm in controls to 5.5–6 μm in mutants. Also note the obvious loss of large caliber axons in the dorsal root (D).
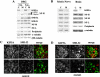
Figure 6.
Accumulation of NF proteins in DRG sensory neuron cell bodies of KIF5A null /KIF5A_flox_; Cre_synapsin_ mutant mice. (A) Western blot analyses of DRGs from the first cohort of 3-wk-old KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice. In the litter shown, control DRGs (C) were pooled from one II/+ and one Cre/+ littermates; mutant DRGs (M) were pooled from two mutant littermates. Note the obvious increase in dephosphorylated NF-H (revealed by the lower band labeled by NF-H, a polyclonal antibody against the COOH terminus of NF-H, and by antibody SMI-32) as well as the increase in NF-M, NF-L, and peripherin. (B) Western blot of brain and sciatic nerve from 3-wk-old KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice. 20 μg of proteins of mutant and control (II/+) littermates was loaded in each lane. Note the clear reduction of KIF5A protein in the mutant. The levels of NFs and peripherin were not significantly changed in the mutant brain and sciatic nerve. (C and D) Immunostaining of DRG sensory neurons of 7.5-mo-old KIF5A_null_/KIF5A_flox_; Cre_synapsin_ mutant mice. Double staining with KIF5A (green) and SMI-32 (recognizes dephosphorylated NF-H, red) was performed on a mutant and a control littermate. (C) Control DRG staining. Bottom panel, higher magnification. (D) Mutant DRG staining, higher magnification in the lower panel. Note the apparent intense SMI-32 staining in some DRG neurons. Bars, 200 μm.
Figure 7.
Analysis of DRG accumulation and axonal transport in second cohort of 3-wk-old KIF5A null /KIF5A_flox_; Cre_synapsin_ mutant mice. (A) Axonal transport of APP, Rab3, synaptophysin, and KIF5C as assessed by Western blotting of proximal and distal segments 6 h after ligation of sciatic nerves from control and mutant animals; unligated controls are from the contralateral unligated nerve. Nerves from each animal were homogenized, and equal amounts of nerve proteins (∼40 μg/lane) were loaded onto SDS-PAGE gels and analyzed by Western blotting. Data from two pairs of mice from different litters are shown. P, proximal side; D, distal side. (B) Analysis of DRG content of mutant and control animals. Approximately 12 μg of DRG lysate from each animal was loaded into each lane. Note increased NF-H and NF-L levels in the mutant DRGs, whereas protein amounts of APP, Rab3, and synaptophysin were not obviously changed. (C) Summary of data from 12 mutant and 12 littermate control animals from five litters. SYP, synaptophysin. As many antibodies as possible were used for reprobing of each filter, but filter lifetime was inconsistent so not all probes could be used on all animals.
Figure 7.
Analysis of DRG accumulation and axonal transport in second cohort of 3-wk-old KIF5A null /KIF5A_flox_; Cre_synapsin_ mutant mice. (A) Axonal transport of APP, Rab3, synaptophysin, and KIF5C as assessed by Western blotting of proximal and distal segments 6 h after ligation of sciatic nerves from control and mutant animals; unligated controls are from the contralateral unligated nerve. Nerves from each animal were homogenized, and equal amounts of nerve proteins (∼40 μg/lane) were loaded onto SDS-PAGE gels and analyzed by Western blotting. Data from two pairs of mice from different litters are shown. P, proximal side; D, distal side. (B) Analysis of DRG content of mutant and control animals. Approximately 12 μg of DRG lysate from each animal was loaded into each lane. Note increased NF-H and NF-L levels in the mutant DRGs, whereas protein amounts of APP, Rab3, and synaptophysin were not obviously changed. (C) Summary of data from 12 mutant and 12 littermate control animals from five litters. SYP, synaptophysin. As many antibodies as possible were used for reprobing of each filter, but filter lifetime was inconsistent so not all probes could be used on all animals.
Figure 7.
Analysis of DRG accumulation and axonal transport in second cohort of 3-wk-old KIF5A null /KIF5A_flox_; Cre_synapsin_ mutant mice. (A) Axonal transport of APP, Rab3, synaptophysin, and KIF5C as assessed by Western blotting of proximal and distal segments 6 h after ligation of sciatic nerves from control and mutant animals; unligated controls are from the contralateral unligated nerve. Nerves from each animal were homogenized, and equal amounts of nerve proteins (∼40 μg/lane) were loaded onto SDS-PAGE gels and analyzed by Western blotting. Data from two pairs of mice from different litters are shown. P, proximal side; D, distal side. (B) Analysis of DRG content of mutant and control animals. Approximately 12 μg of DRG lysate from each animal was loaded into each lane. Note increased NF-H and NF-L levels in the mutant DRGs, whereas protein amounts of APP, Rab3, and synaptophysin were not obviously changed. (C) Summary of data from 12 mutant and 12 littermate control animals from five litters. SYP, synaptophysin. As many antibodies as possible were used for reprobing of each filter, but filter lifetime was inconsistent so not all probes could be used on all animals.
Similar articles
- Slow component of axonal transport is impaired in the proximal axon of transgenic mice with a G93A mutant SOD1 gene.
Sasaki S, Warita H, Abe K, Iwata M. Sasaki S, et al. Acta Neuropathol. 2004 May;107(5):452-60. doi: 10.1007/s00401-004-0838-y. Epub 2004 Mar 17. Acta Neuropathol. 2004. PMID: 15029446 - Neurofilament subunit NF-H modulates axonal diameter by selectively slowing neurofilament transport.
Marszalek JR, Williamson TL, Lee MK, Xu Z, Hoffman PN, Becher MW, Crawford TO, Cleveland DW. Marszalek JR, et al. J Cell Biol. 1996 Nov;135(3):711-24. doi: 10.1083/jcb.135.3.711. J Cell Biol. 1996. PMID: 8909545 Free PMC article. - Defective kinesin heavy chain behavior in mouse kinesin light chain mutants.
Rahman A, Kamal A, Roberts EA, Goldstein LS. Rahman A, et al. J Cell Biol. 1999 Sep 20;146(6):1277-88. doi: 10.1083/jcb.146.6.1277. J Cell Biol. 1999. PMID: 10491391 Free PMC article. - Neurofilaments in health and disease.
Gotow T. Gotow T. Med Electron Microsc. 2000;33(4):173-99. doi: 10.1007/s007950000019. Med Electron Microsc. 2000. PMID: 11810476 Review. - Neuronal intermediate filaments.
Lee MK, Cleveland DW. Lee MK, et al. Annu Rev Neurosci. 1996;19:187-217. doi: 10.1146/annurev.ne.19.030196.001155. Annu Rev Neurosci. 1996. PMID: 8833441 Review.
Cited by
- Differential Co-Expression Analysis of RNA-Seq Data Reveals Novel Potential Biomarkers of Device-Tissue Interaction.
Moore MG, Thompson CH, Reimers MA, Purcell EK. Moore MG, et al. Annu Int Conf IEEE Eng Med Biol Soc. 2022 Jul;2022:3072-3076. doi: 10.1109/EMBC48229.2022.9871437. Annu Int Conf IEEE Eng Med Biol Soc. 2022. PMID: 36085767 Free PMC article. - β-Tubulin mutations that cause severe neuropathies disrupt axonal transport.
Niwa S, Takahashi H, Hirokawa N. Niwa S, et al. EMBO J. 2013 May 15;32(10):1352-64. doi: 10.1038/emboj.2013.59. Epub 2013 Mar 15. EMBO J. 2013. PMID: 23503589 Free PMC article. - KIF5A upregulation in hepatocellular carcinoma: A novel prognostic biomarker associated with unique tumor microenvironment status.
Liu Q, Liu YY, Chen XM, Tao BY, Chen K, Li WM, Xu CT, Shi Y, Li H, Liu HR. Liu Q, et al. Front Oncol. 2023 Jan 6;12:1071722. doi: 10.3389/fonc.2022.1071722. eCollection 2022. Front Oncol. 2023. PMID: 36686769 Free PMC article. - KIF5C deficiency causes abnormal cortical neuronal migration, dendritic branching, and spine morphology in mice.
Li W, Cheng T, Dong X, Chen H, Yang L, Qiu Z, Zhou W. Li W, et al. Pediatr Res. 2022 Oct;92(4):995-1002. doi: 10.1038/s41390-021-01922-8. Epub 2021 Dec 29. Pediatr Res. 2022. PMID: 34966180 - Kinesin-dependent transport of keratin filaments: a unified mechanism for intermediate filament transport.
Robert A, Tian P, Adam SA, Kittisopikul M, Jaqaman K, Goldman RD, Gelfand VI. Robert A, et al. FASEB J. 2019 Jan;33(1):388-399. doi: 10.1096/fj.201800604R. Epub 2018 Jun 26. FASEB J. 2019. PMID: 29944446 Free PMC article.
References
- Bloom, G.S., and S.A. Endow. 1995. Motor proteins 1: kinesins. Protein Profile. 2:1105–1171. - PubMed
- Bloom, G.S., M.C. Gagner, K.K. Pfister, and S.T. Brady. 1988. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 27:3409–3416. - PubMed
- Brady, S.T. 1985. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 317:73–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous