Essential Bacillus subtilis genes - PubMed (original) (raw)
. 2003 Apr 15;100(8):4678-83.
doi: 10.1073/pnas.0730515100. Epub 2003 Apr 7.
S D Ehrlich, A Albertini, G Amati, K K Andersen, M Arnaud, K Asai, S Ashikaga, S Aymerich, P Bessieres, F Boland, S C Brignell, S Bron, K Bunai, J Chapuis, L C Christiansen, A Danchin, M Débarbouille, E Dervyn, E Deuerling, K Devine, S K Devine, O Dreesen, J Errington, S Fillinger, S J Foster, Y Fujita, A Galizzi, R Gardan, C Eschevins, T Fukushima, K Haga, C R Harwood, M Hecker, D Hosoya, M F Hullo, H Kakeshita, D Karamata, Y Kasahara, F Kawamura, K Koga, P Koski, R Kuwana, D Imamura, M Ishimaru, S Ishikawa, I Ishio, D Le Coq, A Masson, C Mauël, R Meima, R P Mellado, A Moir, S Moriya, E Nagakawa, H Nanamiya, S Nakai, P Nygaard, M Ogura, T Ohanan, M O'Reilly, M O'Rourke, Z Pragai, H M Pooley, G Rapoport, J P Rawlins, L A Rivas, C Rivolta, A Sadaie, Y Sadaie, M Sarvas, T Sato, H H Saxild, E Scanlan, W Schumann, J F M L Seegers, J Sekiguchi, A Sekowska, S J Séror, M Simon, P Stragier, R Studer, H Takamatsu, T Tanaka, M Takeuchi, H B Thomaides, V Vagner, J M van Dijl, K Watabe, A Wipat, H Yamamoto, M Yamamoto, Y Yamamoto, K Yamane, K Yata, K Yoshida, H Yoshikawa, U Zuber, N Ogasawara
Affiliations
- PMID: 12682299
- PMCID: PMC153615
- DOI: 10.1073/pnas.0730515100
Essential Bacillus subtilis genes
K Kobayashi et al. Proc Natl Acad Sci U S A. 2003.
Abstract
To estimate the minimal gene set required to sustain bacterial life in nutritious conditions, we carried out a systematic inactivation of Bacillus subtilis genes. Among approximately 4,100 genes of the organism, only 192 were shown to be indispensable by this or previous work. Another 79 genes were predicted to be essential. The vast majority of essential genes were categorized in relatively few domains of cell metabolism, with about half involved in information processing, one-fifth involved in the synthesis of cell envelope and the determination of cell shape and division, and one-tenth related to cell energetics. Only 4% of essential genes encode unknown functions. Most essential genes are present throughout a wide range of Bacteria, and almost 70% can also be found in Archaea and Eucarya. However, essential genes related to cell envelope, shape, division, and respiration tend to be lost from bacteria with small genomes. Unexpectedly, most genes involved in the Embden-Meyerhof-Parnas pathway are essential. Identification of unknown and unexpected essential genes opens research avenues to better understanding of processes that sustain bacterial life.
Figures
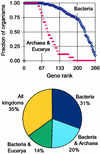
Figure 1
B. subtilis essential gene homologues are widely conserved. (Upper) Genes are ordered by their relative abundance among 54 Bacteria (blue) and 18 Archaea and Eucarya (red). The position (rank) of a gene is shown on abscissa and the fraction of organisms in which a gene is present is shown on the ordinate. (Lower) Fraction of genes present in different kingdoms of life (a gene counted as “all kingdoms” is present in at least one archaeon and one eukaryote, in addition to bacteria, whereas a gene counted as “bacteria” is not present in any archae or eukaryote). The list of genes and organisms is presented in Table 4.
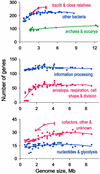
Figure 2
The number of B. subtilis essential gene homologues depends on genome size. (Top) All genes. Bacilli and close relatives denote Bacillus species and other low-G+C Gram-positive bacteria, but not clostridia, mycoplasma, and ureaplasma. (Middle and Bottom) Different bacterial gene categories. Empty red circles in Bottom refer to Bacilli and close relatives, whereas filled red circles refer to other bacteria. Interpolated lines throughout the figure correspond to the best fitting polynomial of the second or the fourth order. The number of genes is: information processing, 136; envelope, respiration, cell shape, and division, 76; cofactors, other, and unknown, 41; and nucleotides and glycolysis, 18.
Figure 3
Phylogenetic profiling of essential genes. The 271 B. subtilis genes were grouped in 266 clusters. Only one gene, yhdL, which encodes a possible anti-sigma protein, had no orthologues in the database and is not presented here. Each line and column corresponds to individual gene and organism, respectively. Presence and absence of a gene is indicated by a black and white square, respectively. The list of genes and organisms is given in Table 5, which is published as supporting information on the PNAS web site and the ordering is described in the text.
References
- Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. - PubMed
- Hutchison C A, Peterson S N, Gill S R, Cline R T, White O, Fraser C M, Smith H O, Venter J C. Science. 1999;286:2165–2169. - PubMed
- Ji Y, Zhang B, Van Horn S F, Warren P, Woodnutt G, Burnham M K, Rosenberg M. Science. 2001;293:2266–2269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials