Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers - PubMed (original) (raw)
. 2003 May;72(5):1171-86.
doi: 10.1086/375120. Epub 2003 Apr 10.
Affiliations
- PMID: 12690579
- PMCID: PMC1180270
- DOI: 10.1086/375120
Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers
Lev A Zhivotovsky et al. Am J Hum Genet. 2003 May.
Abstract
We study data on variation in 52 worldwide populations at 377 autosomal short tandem repeat loci, to infer a demographic history of human populations. Variation at di-, tri-, and tetranucleotide repeat loci is distributed differently, although each class of markers exhibits a decrease of within-population genetic variation in the following order: sub-Saharan Africa, Eurasia, East Asia, Oceania, and America. There is a similar decrease in the frequency of private alleles. With multidimensional scaling, populations belonging to the same major geographic region cluster together, and some regions permit a finer resolution of populations. When a stepwise mutation model is used, a population tree based on TD estimates of divergence time suggests that the branches leading to the present sub-Saharan African populations of hunter-gatherers were the first to diverge from a common ancestral population (approximately 71-142 thousand years ago). The branches corresponding to sub-Saharan African farming populations and those that left Africa diverge next, with subsequent splits of branches for Eurasia, Oceania, East Asia, and America. African hunter-gatherer populations and populations of Oceania and America exhibit no statistically significant signature of growth. The features of population subdivision and growth are discussed in the context of the ancient expansion of modern humans.
Figures
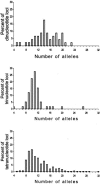
Figure 1
Among-locus distribution of the total number of alleles observed in 1,056 individuals at di-, tri-, and tetranucleotide STRs.
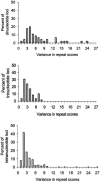
Figure 2
Among-locus distribution of the within-population variance in the repeat scores at di-, tri-, and tetranucleotide STRs. For each STR, the values of the within-population variance were averaged over all 52 populations.
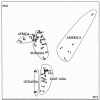
Figure 3
Distribution and clusters of 52 populations from the first two PCs in the multidimensional-scaling plot of _F_ST values at 377 STRs. Most populations are indicated with numbers (see the “Material and Methods” section). The X- and _Y-_axes represent PC1 and PC2, respectively. ⧫ = Mozabite; ◊ = three Middle East samples; □ = Basque, Sardinian, and Orcadian samples; ○ = five other samples from Europe; ▵ = East Asian Altaic-speaking populations; ▴ = two Han populations; * = Kalash; ● = remaining samples.
Figure 4
Multidimensional scaling on the _F_ST values at 377 STRs. A, Separation of sub-Saharan African and American populations in a plot of PC4 versus PC1. B, Separation of Oceania in a plot of PC5 versus PC2 (samples from Africa and America are suppressed).
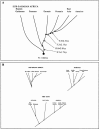
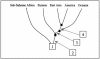
Figure 5
Population tree based on _T_D estimates of divergence time. A. Divergence among major groups. The time estimates are based on 374 STRs (three outlying STRs with tetranucleotide repeats were omitted). Arrows indicate the time (lower bounds, in ky) between adjacent nodes, assuming a generation length of 25 years. B, Schemes of divergence within the major groups, based on the 374 STRs. Time estimates within each continental group were omitted, because they may be biased owing to possible differential gene flows from other groups.
Figure 6
Reduced population tree, showing four separation events (see table 3)
Similar articles
- Human population expansion and microsatellite variation.
Zhivotovsky LA, Bennett L, Bowcock AM, Feldman MW. Zhivotovsky LA, et al. Mol Biol Evol. 2000 May;17(5):757-67. doi: 10.1093/oxfordjournals.molbev.a026354. Mol Biol Evol. 2000. PMID: 10779536 - Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow.
Zhivotovsky LA. Zhivotovsky LA. Mol Biol Evol. 2001 May;18(5):700-9. doi: 10.1093/oxfordjournals.molbev.a003852. Mol Biol Evol. 2001. PMID: 11319254 - Why hunter-gatherer populations do not show signs of pleistocene demographic expansions.
Excoffier L, Schneider S. Excoffier L, et al. Proc Natl Acad Sci U S A. 1999 Sep 14;96(19):10597-602. doi: 10.1073/pnas.96.19.10597. Proc Natl Acad Sci U S A. 1999. PMID: 10485871 Free PMC article. - The human genetic history of East Asia: weaving a complex tapestry.
Stoneking M, Delfin F. Stoneking M, et al. Curr Biol. 2010 Feb 23;20(4):R188-93. doi: 10.1016/j.cub.2009.11.052. Curr Biol. 2010. PMID: 20178766 Review. - Craniometric variation, genetic theory, and modern human origins.
Relethford JH, Harpending HC. Relethford JH, et al. Am J Phys Anthropol. 1994 Nov;95(3):249-70. doi: 10.1002/ajpa.1330950302. Am J Phys Anthropol. 1994. PMID: 7856764 Review.
Cited by
- A population-genetic perspective on the similarities and differences among worldwide human populations.
Rosenberg NA. Rosenberg NA. Hum Biol. 2011 Dec;83(6):659-84. doi: 10.3378/027.083.0601. Hum Biol. 2011. PMID: 22276967 Free PMC article. Review. - A comprehensive analysis of microsatellite diversity in Aboriginal Australians.
Walsh SJ, Mitchell RJ, Watson N, Buckleton JS. Walsh SJ, et al. J Hum Genet. 2007;52(9):712-728. doi: 10.1007/s10038-007-0172-z. Epub 2007 Jul 13. J Hum Genet. 2007. PMID: 17628738 - Whole-genome sequence analysis of a Pan African set of samples reveals archaic gene flow from an extinct basal population of modern humans into sub-Saharan populations.
Lorente-Galdos B, Lao O, Serra-Vidal G, Santpere G, Kuderna LFK, Arauna LR, Fadhlaoui-Zid K, Pimenoff VN, Soodyall H, Zalloua P, Marques-Bonet T, Comas D. Lorente-Galdos B, et al. Genome Biol. 2019 Apr 26;20(1):77. doi: 10.1186/s13059-019-1684-5. Genome Biol. 2019. PMID: 31023378 Free PMC article. - Microsatellites are molecular clocks that support accurate inferences about history.
Sun JX, Mullikin JC, Patterson N, Reich DE. Sun JX, et al. Mol Biol Evol. 2009 May;26(5):1017-27. doi: 10.1093/molbev/msp025. Epub 2009 Feb 12. Mol Biol Evol. 2009. PMID: 19221007 Free PMC article. - Variants of the melanocortin 1 receptor gene (MC1R) and P gene as indicators of the population origin of an individual.
Masui S, Nakatome M, Matoba R. Masui S, et al. Int J Legal Med. 2009 May;123(3):205-11. doi: 10.1007/s00414-008-0289-4. Epub 2008 Oct 7. Int J Legal Med. 2009. PMID: 18839200
References
Electronic-Database Information
- Human Diversity Panel Genotypes, http://research.marshfieldclinic.org/genetics/Freq/FreqInfo.htm (for genotypes used in the present study)
- Human STRP Screening Sets, http://research.marshfieldclinic.org/genetics/sets/combo.html (for Marshfield panel 10)
- Lewis Lab Software, http://lewis.eeb.uconn.edu/lewishome/software.html (for GDA)
References
- Barton, NH, Slatkin M (1986) A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 56:409–416 - PubMed
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457 - PubMed
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, et al (2002) A human genome diversity cell line panel. Science 296:261–262 - PubMed
- Cann RL, Stoneking M, Wilson AC (1987) Mitochondrial DNA and human evolution. Nature 325:31–36 - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM028016/GM/NIGMS NIH HHS/United States
- GM 28016/GM/NIGMS NIH HHS/United States
- P01 GM028428/GM/NIGMS NIH HHS/United States
- R03 TW005540/TW/FIC NIH HHS/United States
- GM 28428/GM/NIGMS NIH HHS/United States
- 1 R03 TW05540/TW/FIC NIH HHS/United States
LinkOut - more resources
Full Text Sources