Regulation of E2F1 by BRCT domain-containing protein TopBP1 - PubMed (original) (raw)
Regulation of E2F1 by BRCT domain-containing protein TopBP1
Kang Liu et al. Mol Cell Biol. 2003 May.
Abstract
The E2F transcription factor integrates cellular signals and coordinates cell cycle progression. Our prior studies demonstrated selective induction and stabilization of E2F1 through ATM-dependent phosphorylation in response to DNA damage. Here we report that DNA topoisomerase IIbeta binding protein 1 (TopBP1) regulates E2F1 during DNA damage. TopBP1 contains eight BRCT (BRCA1 carboxyl-terminal) motifs and upon DNA damage is recruited to stalled replication forks, where it participates in a DNA damage checkpoint. Here we demonstrated an interaction between TopBP1 and E2F1. The interaction depended on the amino terminus of E2F1 and the sixth BRCT domain of TopBP1. It was specific to E2F1 and was not observed in E2F2, E2F3, or E2F4. This interaction was induced by DNA damage and phosphorylation of E2F1 by ATM. Through this interaction, TopBP1 repressed multiple activities of E2F1, including transcriptional activity, induction of S-phase entry, and apoptosis. Furthermore, TopBP1 relocalized E2F1 from diffuse nuclear distribution to discrete punctate nuclear foci, where E2F1 colocalized with TopBP1 and BRCA1. Thus, the specific interaction between TopBP1 and E2F1 during DNA damage inhibits the known E2F1 activities but recruits E2F1 to a BRCA1-containing repair complex, suggesting a direct role of E2F1 in DNA damage checkpoint/repair at stalled replication forks.
Figures
FIG. 1.
Schematic structures of TopBP1 and E2F1. (A) TopBP1 and its six deletion mutants. TopBP1 has eight BRCT domains and a putative nuclear localization signal (NLS). TopBP1CT (encoding the C terminus of TopBP1) was identified in our yeast two-hybrid screen as an E2F1 interactor and contains BRCT6, -7, and -8. HPV16, human papillomavirus type 16. (B) E2F1 and its five deletion mutants. E2F1(1-109), encoding amino acids 1 to 109, was used as a bait for yeast two-hybrid screening. Δ283-358E2F1 encodes an E2F1 mutant with a deletion of the marked box (MB). Act/Rb, activation-retinoblastoma.
FIG. 2.
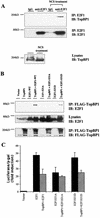
Interaction of E2F1 and TopBP1 in vitro. (A) The interaction of the E2F1 N terminus (amino acids 1 to 109, named E2F1N) and the TopBP1 C terminus (TopBP1CT) in S. cerevisiae Y190 cells was verified by selective growth of transformants on a plate lacking tryptophan, leucine, and histidine and supplemented with 3-amino-1,2,4-triazole. (B) Interaction of TopBP1 and E2F1 in vitro. A GST-TopBP1 pulldown assay was performed. The purified GST or GST-TopBP1 was incubated with E2F1 protein. E2F1 protein was detected by immunoblotting with an anti-E2F1 antibody. GST-TopBP1 and GST were visualized by Ponceau S staining.
FIG. 3.
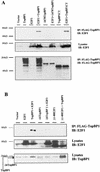
Interaction of TopBP1 and E2F1 in vivo. (A) E2F1 coimmunoprecipitates with TopBP1 or TopBP1CT (containing the sixth, seventh, and eighth carboxyl BRCT motifs) but not with mutants of TopBP1 in which the sixth, seventh, and eighth BRCT motifs were deleted (Δ678TopBP1). 293T cells were transfected with expression plasmids as indicated at the top, and lysates were immunoprecipitated (IP) with anti-Flag-conjugated agarose beads and immunoblotted (IB) as indicated. A portion of the cell lysates before immunoprecipitation was probed with an anti-E2F1 antibody (C20), which detected endogenous E2F1 and transfected E2F1. (B) Mapping the interacting regions of TopBP1 and E2F1. A deletion of the E2F1 N terminus or the sixth BRCT domain of TopBP1 eliminated or greatly reduced E2F1-TopBP1 interaction in the in vivo coimmunoprecipitation assay described for panel A. Total lysates were immunoblotted with E2F1 and TopBP1 antibodies that recognized both endogenous and transfected proteins (bottom two panels). (C) The sixth BRCT domain of TopBP1 binds E2F1. The Flag-tagged BRCT6 motif of TopBP1 was able to coimmunoprecipitate with E2F1 in vivo in 293T cells. The immunoprecipitation was performed as described for panel A.
FIG. 3.
Interaction of TopBP1 and E2F1 in vivo. (A) E2F1 coimmunoprecipitates with TopBP1 or TopBP1CT (containing the sixth, seventh, and eighth carboxyl BRCT motifs) but not with mutants of TopBP1 in which the sixth, seventh, and eighth BRCT motifs were deleted (Δ678TopBP1). 293T cells were transfected with expression plasmids as indicated at the top, and lysates were immunoprecipitated (IP) with anti-Flag-conjugated agarose beads and immunoblotted (IB) as indicated. A portion of the cell lysates before immunoprecipitation was probed with an anti-E2F1 antibody (C20), which detected endogenous E2F1 and transfected E2F1. (B) Mapping the interacting regions of TopBP1 and E2F1. A deletion of the E2F1 N terminus or the sixth BRCT domain of TopBP1 eliminated or greatly reduced E2F1-TopBP1 interaction in the in vivo coimmunoprecipitation assay described for panel A. Total lysates were immunoblotted with E2F1 and TopBP1 antibodies that recognized both endogenous and transfected proteins (bottom two panels). (C) The sixth BRCT domain of TopBP1 binds E2F1. The Flag-tagged BRCT6 motif of TopBP1 was able to coimmunoprecipitate with E2F1 in vivo in 293T cells. The immunoprecipitation was performed as described for panel A.
FIG. 4.
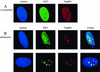
DNA damage induces E2F1 foci. (A) Human foreskin fibroblasts were fixed and immunostained with specific antibodies against E2F1 (polyclonal) and TopBP1 (monoclonal), followed by fluorescein isothiocyanate-conjugated anti-rabbit IgG and Texas Red X-conjugated anti-mouse IgG, respectively. Nuclei were stained with Hoechst 33258. (B) HFF were treated with adriamycin (1 μM) for 6 h, and cells were immunostained as for panel A. Two representative cells are shown.
FIG.5.
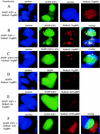
TopBP1 recruits E2F1 to nuclear foci. 293 cells were transfected with expression plasmids as indicated on the left. Cells were fixed, and nuclei were stained with Hoechst 33258. (A) When expressed alone, EGFP-E2F1 formed a homogenous nuclear pattern and HcRed1-TopBP1 formed punctate nuclear foci. (B) Upon coexpression with TopBP1, EGFP-E2F1 formed foci and colocalized with TopBP1. (C) HcRed1-TopBP1 also induced EGFP-E2F1(1-83) to form foci with TopBP1. (D) TopBP1 did not relocalize EGFP to foci. (E) The ability to recruit E2F1 to the foci was greatly impaired in the Δ6TopBP1 mutant. (F) Δ78TopBP1, lacking the nuclear localization signal in the carboxyl terminus, was mainly located in the cytoplasm.
FIG. 6.
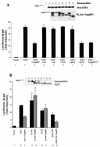
TopBP1 regulates E2F1 transcriptional activity. (A) E2F1 activity was measured with a p14ARF promoter-luciferase activity assay in 293T cells. Luciferase activity of transfected E2F1 is given as induction relative to that of the empty vector control. Each sample was tested in triplicate, and the experiments were repeated multiple times with consistent results. E2F1 activity was assayed in the presence of TopBP1 and six different TopBP1 mutants. A portion of the cell lysates was immunoblotted with antibody for HA or immunoprecipitated with anti-Flag-agarose, followed by anti-Flag immunoblotting. (B) The E2F1 N terminus is required for E2F1 activity to be repressed by TopBP1. Cotransfections of TopBP1 with E2F1 and its three deletion mutants were performed, and cells were prepared for the luciferase assay as described for panel A. Also shown is the immunoblot probed with anti-E2F1 (C20), which detects endogenous E2F1 and transfected wild-type or mutant E2F1. (C) E2F1 transcriptional activity was assayed as for A. E2F1 expression plasmid (1 μg) was cotransfected with increasing amounts of TopBP1 plasmid (0 to 20 μg). The total amount of DNA transfected for each sample was made equivalent with pcDNA3 vector plasmid DNA. The values indicated are the induction relative to the empty vector control. (D) E2F1 activity was measured with a DNA polymerase α p68 promoter-luciferase activity assay. The two plasmids containing DNA polymerase α promoter-luciferase, pKL12 and pKL12 E2FAB, were used as an additional indicator of E2F activity. E2F binding sites are preserved (pKL12) or deleted (pKL12 E2FAB) in these plasmids.
FIG. 6.
TopBP1 regulates E2F1 transcriptional activity. (A) E2F1 activity was measured with a p14ARF promoter-luciferase activity assay in 293T cells. Luciferase activity of transfected E2F1 is given as induction relative to that of the empty vector control. Each sample was tested in triplicate, and the experiments were repeated multiple times with consistent results. E2F1 activity was assayed in the presence of TopBP1 and six different TopBP1 mutants. A portion of the cell lysates was immunoblotted with antibody for HA or immunoprecipitated with anti-Flag-agarose, followed by anti-Flag immunoblotting. (B) The E2F1 N terminus is required for E2F1 activity to be repressed by TopBP1. Cotransfections of TopBP1 with E2F1 and its three deletion mutants were performed, and cells were prepared for the luciferase assay as described for panel A. Also shown is the immunoblot probed with anti-E2F1 (C20), which detects endogenous E2F1 and transfected wild-type or mutant E2F1. (C) E2F1 transcriptional activity was assayed as for A. E2F1 expression plasmid (1 μg) was cotransfected with increasing amounts of TopBP1 plasmid (0 to 20 μg). The total amount of DNA transfected for each sample was made equivalent with pcDNA3 vector plasmid DNA. The values indicated are the induction relative to the empty vector control. (D) E2F1 activity was measured with a DNA polymerase α p68 promoter-luciferase activity assay. The two plasmids containing DNA polymerase α promoter-luciferase, pKL12 and pKL12 E2FAB, were used as an additional indicator of E2F activity. E2F binding sites are preserved (pKL12) or deleted (pKL12 E2FAB) in these plasmids.
FIG.7.
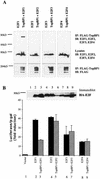
Interaction between E2F1 and TopBP1 during DNA damage. (A) Interaction between endogenous E2F1 and TopBP1. 293T cells were untreated or treated with the radiomimetic agent neocarzinostatin (NCS) at 300 ng/ml for 3 h. Lysates were immunoprecipitated with normal rabbit IgG or rabbit anti-E2F1 antibody (C20) and immunoblotted as indicated. Coimmunoprecipitation of endogenous TopBP1 and E2F1 was detected only in NCS-treated cells. An aliquot of lysates was probed with TopBP1 antibody to detect endogenous TopBP1 (lower panel). (B) TopBP1 interacts with E2F1 and E2F1-S31D but not E2F1-S31A. Flag-tagged TopBP1 was immunoprecipitated from the cotransfected cells, and the bound E2F1 protein was detected by immunoblotting as described for Fig. 3A. Total lysates were probed with anti-E2F1 (C20), which detects endogenous as well as overexpressed E2F1 (middle panel). (C) TopBP1 represses the transcriptional activity of E2F1 and E2F1-S31D but not E2F1-S31A. The E2F1 activity assay with the p14ARF promoter-luciferase construct was performed as described for Fig. 6A.
FIG. 8.
E2F/TopBP1 interaction is specific to E2F1. (A) Specific TopBP1-E2F1 interaction in vivo. E2F1, E2F2, E2F3, and E2F4 were cotransfected with Flag-tagged TopBP1 in 293T cells as described for Fig. 3A. TopBP1 was immunoprecipitated, and the precipitates were probed with an appropriate anti-E2F antibody. No interaction was observed between TopBP1 and E2F2, E2F3, or E2F4. (B) Specificity of TopBP1 regulation of E2F1 transcriptional activity. E2F activity assay with the p14ARF promoter-luciferase construct was performed as described for Fig. 6A. All E2Fs were HA tagged. A portion of the cell lysate was immunoblotted with an anti-HA antibody. TopBP1 has no significant effect on the transcriptional activities of E2F2, E2F3, or E2F4.
FIG. 9.
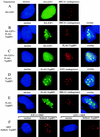
TopBP1 recruits E2F1 to BRCA1-containing complex. (A to E) 293 cells were transfected as indicated on the left. Cells were fixed and immunostained with specific antibodies against E2F1, BRCA1, or TopBP1 and fluorescein isothiocyanate- or Texas Red X-conjugated secondary antibodies. Nuclei were stained with Hoechst 33258. HA-E2F1 was distributed as a homogenous nuclear pattern (A), but was relocalized to punctate nuclear foci upon coexpression with Flag-TopBP1, where it colocalized with BRCA1 (B) and TopBP1 (C). Furthermore, both endogenous E2F1 (D) and BRCA1 (E) colocalized with TopBP1 upon overexpression of Flag-TopBP1. (F) Primary mouse embryo fibroblasts (MEFs) prepared from E2F1+/+ and E2F1−/− sibling embryos were transfected with pHcRed1-TopBP1. Nuclei were stained with Hoechst 33258. TopBP1 formed foci in both cells.
FIG. 10.
TopBP1 regulates E2F1 function. (A) TopBP1 inhibits E2F1 S-phase activation. REF52 cells were serum starved for 48 h and then infected with AdE2F1 and/or AdTopBP1 at a multiplicity of infection of 400. Some cells infected with AdCMV or AdTopBP1 were stimulated with 15% serum. BrdU labeling was performed between 19 and 40 h after infection. Incorporated BrdU was detected with anti-BrdU antibody and Texas Red-conjugated secondary antibody. Nuclei were stained with Hoechst 33258. About 2,500 nuclei were scored on each sample for BrdU incorporation by microscopy. The experiments were performed multiple times with similar results. One representative result is shown. (B) TopBP1 inhibits E2F1-induced apoptosis. Similar infections were performed in serum-starved REF52 cells as for panel A. Apoptosis was assayed 4 days after infection, and apoptotic cells were counted by propidium iodide-flow cytometry. The percentage of apoptosis is expressed relative to the total number of cells counted.
Similar articles
- TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival.
Liu K, Luo Y, Lin FT, Lin WC. Liu K, et al. Genes Dev. 2004 Mar 15;18(6):673-86. doi: 10.1101/gad.1180204. Genes Dev. 2004. PMID: 15075294 Free PMC article. - A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival.
Yamane K, Wu X, Chen J. Yamane K, et al. Mol Cell Biol. 2002 Jan;22(2):555-66. doi: 10.1128/MCB.22.2.555-566.2002. Mol Cell Biol. 2002. PMID: 11756551 Free PMC article. - Divergent siblings: E2F2 and E2F4 but not E2F1 and E2F3 induce DNA synthesis in cardiomyocytes without activation of apoptosis.
Ebelt H, Hufnagel N, Neuhaus P, Neuhaus H, Gajawada P, Simm A, Müller-Werdan U, Werdan K, Braun T. Ebelt H, et al. Circ Res. 2005 Mar 18;96(5):509-17. doi: 10.1161/01.RES.0000159705.17322.57. Epub 2005 Feb 17. Circ Res. 2005. PMID: 15718499 - A new role for E2F-1 in checkpoint control.
Stevens C, La Thangue NB. Stevens C, et al. Cell Cycle. 2003 Sep-Oct;2(5):435-7. Cell Cycle. 2003. PMID: 12963836 Review.
Cited by
- RNF126 promotes homologous recombination via regulation of E2F1-mediated BRCA1 expression.
Wang Y, Deng O, Feng Z, Du Z, Xiong X, Lai J, Yang X, Xu M, Wang H, Taylor D, Yan C, Chen C, Difeo A, Ma Z, Zhang J. Wang Y, et al. Oncogene. 2016 Mar 17;35(11):1363-72. doi: 10.1038/onc.2015.198. Epub 2015 Aug 3. Oncogene. 2016. PMID: 26234677 Free PMC article. - Life and death decisions by the E2F transcription factors.
Iaquinta PJ, Lees JA. Iaquinta PJ, et al. Curr Opin Cell Biol. 2007 Dec;19(6):649-57. doi: 10.1016/j.ceb.2007.10.006. Epub 2007 Nov 26. Curr Opin Cell Biol. 2007. PMID: 18032011 Free PMC article. Review. - GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage.
Guo R, Chen J, Mitchell DL, Johnson DG. Guo R, et al. Nucleic Acids Res. 2011 Mar;39(4):1390-7. doi: 10.1093/nar/gkq983. Epub 2010 Oct 23. Nucleic Acids Res. 2011. PMID: 20972224 Free PMC article. - E2F1 plays a direct role in Rb stabilization and p53-independent tumor suppression.
Palacios G, Talos F, Nemajerova A, Moll UM, Petrenko O. Palacios G, et al. Cell Cycle. 2008 Jun 15;7(12):1776-81. doi: 10.4161/cc.7.12.6030. Epub 2008 Jun 30. Cell Cycle. 2008. PMID: 18583939 Free PMC article. - RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1.
Vélez-Cruz R, Manickavinayaham S, Biswas AK, Clary RW, Premkumar T, Cole F, Johnson DG. Vélez-Cruz R, et al. Genes Dev. 2016 Nov 15;30(22):2500-2512. doi: 10.1101/gad.288282.116. Genes Dev. 2016. PMID: 27940962 Free PMC article.
References
- Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. - PubMed
- Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. - PubMed
- Bochar, D. A., L. Wang, H. Beniya, A. Kinev, Y. Xue, W. S. Lane, W. Wang, F. Kashanchi, and R. Shiekhattar. 2000. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102:257-265. - PubMed
- Boner, W., E. R. Taylor, E. Tsirimonaki, K. Yamane, M. S. Campo, and I. M. Morgan. 2002. A functional interaction between the human papillomavirus type 16 transcription/replication factor E2 and the DNA damage response protein TopBP1. J. Biol. Chem. 4:4. - PubMed
- Bork, P., K. Hofmann, P. Bucher, A. F. Neuwald, S. F. Altschul, and E. V. Koonin. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11:68-76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous