MatchMiner: a tool for batch navigation among gene and gene product identifiers - PubMed (original) (raw)
MatchMiner: a tool for batch navigation among gene and gene product identifiers
Kimberly J Bussey et al. Genome Biol. 2003.
Abstract
MatchMiner is a freely available program package for batch navigation among gene and gene product identifier types commonly encountered in microarray studies and other forms of 'omic' research. The user inputs a list of gene identifiers and then uses the Merge function to find the overlap with a second list of identifiers of either the same or a different type or uses the LookUp function to find corresponding identifiers.
Figures
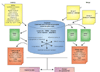
Figure 1
Information Flow in MatchMiner. Input identifier lists are first translated into unique internal gene indices to form a translation table. The translation table is then either converted into another set of identifiers using the LookUp function or compared with another such table using the Merge function to generate a report showing the intersection of two separate identifier lists. The resulting output can be displayed as HTML or else saved as text for import into other programs.
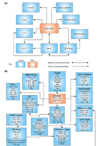
Figure 2
Database relational table schema for MatchMiner. (a) Logical database representation. Data are incorporated from the UCSC Human Genome Build, LocusLink, UniGene, OMIM, and the Affymetrix annotation sets for HU95 and HU133 chips. Each candidate gene is assigned a gene index in the GeneIdx table. These gene indexes are used as keys for all of the MatchMiner operations. The number of many-to-many relationships in the model illustrates the complexity of the data. (b) Physical representation of the database. The implementation currently includes 14 tables with about 12 million rows.
Figure 3
Associating FISH-mapped BACs with genes. Schematic view of FISH-mapped BACs from 1p36.33 near the PITSLRE kinase genes (UCSC Genome Browser, June 2002 freeze). Note that a single BAC can encompass one or more genes. In MatchMiner, the FISH-mapped BAC table from UCSC is imported, and chromosomal positions are read from the table for comparison with the transcriptional start positions of UCSC 'Known Genes'. If a transcriptional start is contained within the bounds of a BAC, that BAC is associated with the corresponding gene index. Thus, a BAC containing several genes will be associated with each of those genes.
Similar articles
- Graphical representation of ribosomal RNA probe accessibility data using ARB software package.
Kumar Y, Westram R, Behrens S, Fuchs B, Glöckner FO, Amann R, Meier H, Ludwig W. Kumar Y, et al. BMC Bioinformatics. 2005 Mar 21;6:61. doi: 10.1186/1471-2105-6-61. BMC Bioinformatics. 2005. PMID: 15777482 Free PMC article. - MILANO--custom annotation of microarray results using automatic literature searches.
Rubinstein R, Simon I. Rubinstein R, et al. BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article. - CrossHybDetector: detection of cross-hybridization events in DNA microarray experiments.
Uva P, de Rinaldis E. Uva P, et al. BMC Bioinformatics. 2008 Nov 17;9:485. doi: 10.1186/1471-2105-9-485. BMC Bioinformatics. 2008. PMID: 19014642 Free PMC article. - Interpreting microarray results with gene ontology and MeSH.
Osborne JD, Zhu LJ, Lin SM, Kibbe WA. Osborne JD, et al. Methods Mol Biol. 2007;377:223-42. doi: 10.1007/978-1-59745-390-5_14. Methods Mol Biol. 2007. PMID: 17634620 Review. - Microarray analysis: genome-scale hypothesis scanning.
Gibson G. Gibson G. PLoS Biol. 2003 Oct;1(1):E15. doi: 10.1371/journal.pbio.0000015. Epub 2003 Oct 13. PLoS Biol. 2003. PMID: 14551912 Free PMC article. Review.
Cited by
- A decision theory paradigm for evaluating identifier mapping and filtering methods using data integration.
Day RS, McDade KK. Day RS, et al. BMC Bioinformatics. 2013 Jul 15;14:223. doi: 10.1186/1471-2105-14-223. BMC Bioinformatics. 2013. PMID: 23855655 Free PMC article. - MicroRNA-9 modified bone marrow-derived mesenchymal stem cells (BMSCs) repair severe acute pancreatitis (SAP) via inducing angiogenesis in rats.
Qian D, Song G, Ma Z, Wang G, Jin L, Hu M, Song Z, Wang X. Qian D, et al. Stem Cell Res Ther. 2018 Oct 25;9(1):282. doi: 10.1186/s13287-018-1022-y. Stem Cell Res Ther. 2018. PMID: 30359310 Free PMC article. Retracted. - B.E.A.R. GeneInfo: a tool for identifying gene-related biomedical publications through user modifiable queries.
Zhou G, Wen X, Liu H, Schlicht MJ, Hessner MJ, Tonellato PJ, Datta MW. Zhou G, et al. BMC Bioinformatics. 2004 Apr 29;5:46. doi: 10.1186/1471-2105-5-46. BMC Bioinformatics. 2004. PMID: 15117422 Free PMC article. - IDconverter and IDClight: conversion and annotation of gene and protein IDs.
Alibés A, Yankilevich P, Cañada A, Díaz-Uriarte R. Alibés A, et al. BMC Bioinformatics. 2007 Jan 10;8:9. doi: 10.1186/1471-2105-8-9. BMC Bioinformatics. 2007. PMID: 17214880 Free PMC article. - Molecular profiling for precision cancer therapies.
Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Malone ER, et al. Genome Med. 2020 Jan 14;12(1):8. doi: 10.1186/s13073-019-0703-1. Genome Med. 2020. PMID: 31937368 Free PMC article. Review.
References
- Weinstein JN. Fishing expeditions. Science. 1998;282:628–629. - PubMed
- Weinstein JN. 'Omic' and hypothesis-driven research in the molecular pharmacology of cancer. Curr Opin Pharmacol. 2002;2:361–365. - PubMed
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources