RNA editing in hornwort chloroplasts makes more than half the genes functional - PubMed (original) (raw)
RNA editing in hornwort chloroplasts makes more than half the genes functional
Masanori Kugita et al. Nucleic Acids Res. 2003.
Abstract
RNA editing in chloroplasts alters the RNA sequence by converting C-to-U or U-to-C at a specific site. During the study of the complete nucleotide sequence of the chloroplast genome from the hornwort Anthoceros formosae, RNA editing events have been systematically investigated. A total of 509 C-to-U and 433 U-to-C conversions are identified in the transcripts of 68 genes and eight ORFs. No RNA editing is seen in any of the rRNA but one tRNA suffered a C-to-U conversion at an anticodon. All nonsense codons in 52 protein-coding genes and seven ORFs are removed in the transcripts by U-to-C conversions, and five initiation and three termination codons are created by C-to-U conversions. RNA editing in intron sequence suggests that editing can precede intercistronic processing. The sequence complementary to the edited site is proposed as a distant cis-recognition element.
Figures
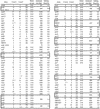
Figure 1
Number of editing sites in the chloroplast of A.formosae. The number of RNA editing sites converting C-to-U and U-to-C is shown. The number of those converting nonsense codons into sense codons is shown in parentheses. Editing sites, which do not alter the coding amino acid are shown as silent (in) and those found outside of the coding region as (out). Editing frequency shows percentage of editing sites per analyzed size (bp). Editing sites on co-transcripts are shown in boxes. *Containing the creation of an initiation codon. (†) Containing the creation of a termination codon.
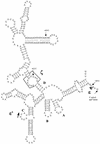
Figure 2
Schematic representation of the predicted secondary structure of domain I in the group II intron of Anthoceros atpF. Sub-domains A, B, C and D together with ε, ε’ and ζ sites are shown. Edited sites 149, 419 and 612 positions from the translation start, and the 5′ end of the intron are shown with arrows.
Similar articles
- Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Tobacco Product Use and Associated Factors Among Middle and High School Students - National Youth Tobacco Survey, United States, 2021.
Gentzke AS, Wang TW, Cornelius M, Park-Lee E, Ren C, Sawdey MD, Cullen KA, Loretan C, Jamal A, Homa DM. Gentzke AS, et al. MMWR Surveill Summ. 2022 Mar 11;71(5):1-29. doi: 10.15585/mmwr.ss7105a1. MMWR Surveill Summ. 2022. PMID: 35271557 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Design of an international, phase IV, open-label study of simoctocog alfa in women/girls with hemophilia A undergoing surgery (NuDIMENSION).
Marquardt N, Langer F, Holstein K, Álvarez Román MT, Núñez Vázquez R, Miljić P, Drillaud N, Ardillon L, Lehtinen AE, Santoro RC, Napolitano M, Siragusa S, Gidley G, Jansen M, Knaub S, Oldenburg J. Marquardt N, et al. Ther Adv Hematol. 2024 Dec 1;15:20406207241300040. doi: 10.1177/20406207241300040. eCollection 2024. Ther Adv Hematol. 2024. PMID: 39624518 Free PMC article. - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- Nuclear DYW-type PPR gene families diversify with increasing RNA editing frequencies in liverwort and moss mitochondria.
Rüdinger M, Volkmar U, Lenz H, Groth-Malonek M, Knoop V. Rüdinger M, et al. J Mol Evol. 2012 Feb;74(1-2):37-51. doi: 10.1007/s00239-012-9486-3. J Mol Evol. 2012. PMID: 22302222 - Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes.
Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. Saski C, et al. Theor Appl Genet. 2007 Aug;115(4):571-90. doi: 10.1007/s00122-007-0567-4. Epub 2007 May 30. Theor Appl Genet. 2007. PMID: 17534593 Free PMC article. - LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis.
Cai W, Ji D, Peng L, Guo J, Ma J, Zou M, Lu C, Zhang L. Cai W, et al. Plant Physiol. 2009 Jul;150(3):1260-71. doi: 10.1104/pp.109.136812. Epub 2009 May 15. Plant Physiol. 2009. PMID: 19448041 Free PMC article.
References
- Covello P.S. and Gray,M.W. (1989) RNA editing in plant mitochondria. Nature, 341, 662–666. - PubMed
- Gualberto J.M., Lamattina,L., Bonnard,G., Weil,J.H. and Grienenberger,J.M. (1989) RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature, 341, 660–662. - PubMed
- Hiesel R., Wissinger,B., Schuster,W. and Brennicke,A. (1989) RNA editing in plant mitochondria. Science, 246, 1632–1634. - PubMed
- Hoch B., Maier,R.M., Appel,K., Igloi,G.L. and Koessel,H. (1991) Editing of a chloroplast mRNA by creation of an initiation codon. Nature, 353, 178–180. - PubMed
- Yu W. and Schuster,W. (1995) Evidence for a site-specific cytidine deamination reaction involved in C to U RNA editing of plant mitochondria. J. Biol. Chem., 270, 18227–18233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases