Independent functions of yeast Pcf11p in pre-mRNA 3' end processing and in transcription termination - PubMed (original) (raw)
Independent functions of yeast Pcf11p in pre-mRNA 3' end processing and in transcription termination
Martin Sadowski et al. EMBO J. 2003.
Abstract
Pcf11p, an essential subunit of the yeast cleavage factor IA, is required for pre-mRNA 3' end processing, binds to the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNAP II) and is involved in transcription termination. We show that the conserved CTD interaction domain (CID) of Pcf11p is essential for cell viability. Interestingly, the CTD binding and 3' end processing activities of Pcf11p can be functionally uncoupled from each other and provided by distinct Pcf11p fragments in trans. Impaired CTD binding did not affect the 3' end processing activity of Pcf11p and a deficiency of Pcf11p in 3' end processing did not prevent CTD binding. Transcriptional run-on analysis with the CYC1 gene revealed that loss of cleavage activity did not correlate with a defect in transcription termination, whereas loss of CTD binding did. We conclude that Pcf11p is a bifunctional protein and that transcript cleavage is not an obligatory step prior to RNAP II termination.
Figures
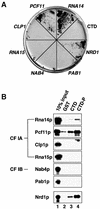
Fig. 1. Interactions of CF IA subunits with the CTD. (A) Two-hybrid interactions of RNA14 and PCF11 with the CTD of RNAP II activate HIS3 reporter gene expression. Y190 cells were cotransformed with pAS2ΔΔ-CTD (CTD fused to the GAL4 DNA-binding domain) and pACT2 (GAL4 activation domain) containing the open reading frames encoding CF IA (RNA14, PCF11, CLP1 and RNA15), CF IB (NAB4), PAB1 and NRD1. HIS3 reporter gene expression was assayed by growth on medium lacking histidine. (B) GST (lane 2), GST–CTD (lane 3) or phosphorylated GST–CTD-P (lane 4) bound to glutathione–Sepharose 4B was incubated with in vitro translated 35S- labelled subunits of CF IA (Rna14p, Pcf11p, Clp1p and Rna15p), CF IB (Nab4p) and Pab1p as indicated on the left of each panel. Bound proteins were separated by SDS–PAGE and visualized by autoradiography. Input (lane 1) shows 10% of in vitro translation reactions used in the binding reaction. The genuine CTD binding protein Nrd1p was used as a positive control.
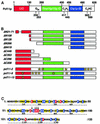
Fig. 2. Pcf11p contains a conserved CTD interaction domain. (A) Schematic representation of the domain organization of Pcf11p. The CTD interaction domain is indicated as CID, the segment of 20 consecutive glutamines as Q20, the Rna14p/Rna15p interaction domain as Rna14p/Rna15p ID, the Clp1p-interaction domain as Clp1p ID, and the two zinc-finger motifs as C2H2 and C2HC, respectively. The numbers correspond to the amino acid sequence of Pcf11p. (B) Schematic representation of Pcf11p mutants used in this study. The numbers and letters indicate the length of the amino acid sequence deleted either at the N- or C-terminus. (C) Sequence of the conserved CID of Pcf11p. The amino acid conservation of the CID of Pcf11p was calculated by sequence alignments of 13 known and putative CTD binding proteins containing the CID (results not shown). Invariant residues are in red boxes (100% conservation), blue boxes indicate conserved and similar amino acids that are present in 70–100% of the aligned sequences and yellow boxes indicate conserved and similar residues common in 30–70% of the aligned sequences. Point mutations in pcf11-9 (A66D) and pcf11-13 (D68A, S69A, I70A) changing the CID sequence are marked by arrows.
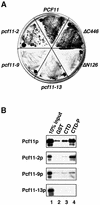
Fig. 3. The CID is responsible for CTD binding. (A) The N-terminal 180 amino acids comprising the CID of Pcf11p are sufficient for a two-hybrid interaction with the CTD. Y190 cells were cotransformed with pAS2ΔΔ-CTD and the pACT2 constructs encoding the open reading frames: PCF11, Δ_N126_, Δ_C446_, pcf11-2, pcf11-9 and pcf11-13. HIS3 reporter gene expression was assayed by growth on medium lacking histidine. (B) Analysis of the CTD binding ability of Pcf11p mutants by GST pull-down experiments. GST (lane 2), GST–CTD (lane 3) or GST–CTD-P (lane 4) bound to glutathione–Sepharose 4B was incubated with in vitro translated 35S-labelled proteins as indicated on the left of the panel. Bound proteins were separated by SDS–PAGE and visualized by autoradiography. Input (lane 1) shows 10% of in vitro translation reactions included in the binding reaction.
Fig. 4. The CID of Pcf11p is essential for cell viability. (A) pcf11-13 complements a complete chromosomal deletion of PCF11. Strain NA53 was transformed with plasmid-encoded PCF11, Δ_C208_ or the CID mutants pcf11-13, Δ_N21-71_, Δ_N108_, Δ_N126_ and Δ_N266_. Transformants were forced to lose the URA3_-marked plasmid pFL38-PCF11 on 5-FOA plates at 23°C. (B) Mutations in pcf11-13 cause temperature sensitivity. Serial dilutions of PCF11 and pcf11-13 cells were incubated at 30 and 37°C. (C) Cross-complementation of the pcf11-2 allele in trans. The pcf11-2 strain was transformed with plasmids encoding PCF11, pcf11-2, the CID mutants pcf11-13, Δ_N21-71, Δ_N108_, Δ_N126_ and Δ_N266_ or empty plasmid. Transformants were grown at restrictive conditions (37°C) for the pcf11-2 allele. (D) Cross-complementation of the pcf11-13 allele in trans. The pcf11-13 strain was transformed with plasmid-encoded PCF11, Δ_N21-71_, Δ_N108_, Δ_N126_, Δ_C208_, Δ_C296_, Δ_C353_ and Δ_C446_. Transformants were grown at restrictive growth conditions (37°C) for the pcf11-13 allele. (E) Western blot analysis of total protein extracts obtained from pcf11-13 (lanes 1–5) and pcf11-2 strains (lanes 6–9) that also carry plasmid-borne N- or C-terminally truncated deletions of Pcf11p as indicated. All deletion proteins (indicated by arrows) were N-terminal in-frame fusions with two IgG binding domains. Pcf11-13p (also containing two N-terminal IgG binding domains) and Pcf11-2 proteins are indicated by asterisks. The blot was consecutively decorated with a polyclonal anti-Pcf11p serum and anti-rabbit IgG–horseradish peroxidase conjugate.
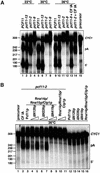
Fig. 5. CTD binding deficiency of Pcf11p does not affect its function in cleavage and polyadenylation in vitro. (A) Mutations in the pcf11-13 allele do not affect cleavage and polyadenylation. 32P-labelled CYC1 RNA (lane 15) was assayed at 23°C (lanes 1–4), 30°C (lanes 5–8) or 36°C (lanes 9–14) under standard reaction conditions in extracts made from wild-type PCF11 (lanes 1, 5 and 9), pcf11-13 (lanes 2, 6 and 10), pcf11-2 (lanes 3, 7, 11 and 14) or pcf11-9 cells (lanes 4, 8, 12 and 13). Restoration of cleavage and polyadenylation in pcf11-2 and pcf11-9 extract at restrictive temperature (36°C) was examined by addition of purified CF IA (lanes 13 and 14). (B) The CID is not necessary for 3′ end processing. Restoration of cleavage and polyadenylation of pcf11-2 extract at restrictive temperature (36°C) was examined by addition of purified CF IA (lane 2) or recombinant GST fusion proteins of Pcf11p (100 ng, lane 3), ΔN126p (50 and 100 ng, lanes 4 and 5), ΔN266p (50 and 100 ng, lanes 6 and 7) and ΔN419p (50 and 100 ng, lanes 8 and 9). Before addition to the pcf11-2 extract, proteins were incubated for 10 min with a combination of recombinant Rna14p, Clp1p and Rna15p (each 100 ng, lanes 3–11) to assemble CF IA. Recombinant proteins were also controlled for endogenous activity (lanes 12–16). Reaction products were visualized by autoradiography after electrophoresis in a gel of 6% polyacrylamide and 8.3 M urea. The positions of the full-length transcript (CYC1), the 5′-cleavage product (5′) and the polyadenylation product (pA) are marked on the right. The length in nucleotides of marker bands (_Hpa_II-digested pBR322 DNA) is indicated on the left.
Fig. 6. Mutations in the CTD-binding domain impair correct transcription termination. (A) Schematic diagram of plasmid pUG_CYC1_ showing the arrangement of M13 probes relative to the CYC1 poly(A) site (position 506). (B) Transcriptional run-on analyses performed in cells transformed with pUG_CYC1_ under permissive growth conditions (23°C) and after shifting to restrictive temperature (37°C) for 60 min (45 min in the case of pcf11-13 + Δ_C208_) as indicated. The numbers at the top of the panel correspond to the probes. Lane M marks the M13 probe used as a background hybridization control. Hybridization of transcripts to the actin probe (lane A) and RNAP III transcripts to the tRNA probe (lane t) are shown. Quantitative analyses of transcriptional run-on profiles are shown on the right of each panel. PhosphorImager quantitation was performed with IMAGEQUANT software. The M13 background was subtracted from each probe, and the results were normalized to probe 1, which was fixed at 100%. Each experiment was performed at least three times, and average values are presented.
Similar articles
- Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors.
Meinhart A, Cramer P. Meinhart A, et al. Nature. 2004 Jul 8;430(6996):223-6. doi: 10.1038/nature02679. Nature. 2004. PMID: 15241417 - Distinct roles of two Yth1p domains in 3'-end cleavage and polyadenylation of yeast pre-mRNAs.
Barabino SM, Ohnacker M, Keller W. Barabino SM, et al. EMBO J. 2000 Jul 17;19(14):3778-87. doi: 10.1093/emboj/19.14.3778. EMBO J. 2000. PMID: 10899131 Free PMC article. - Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors.
de Vries H, Rüegsegger U, Hübner W, Friedlein A, Langen H, Keller W. de Vries H, et al. EMBO J. 2000 Nov 1;19(21):5895-904. doi: 10.1093/emboj/19.21.5895. EMBO J. 2000. PMID: 11060040 Free PMC article. - [C-terminal domain (CTD) of the subunit Rpb1 of nuclear RNA polymerase II and its role in the transcription cycle].
Sobennikova MV, Shematorova EK, Shpakovskiĭ GV. Sobennikova MV, et al. Mol Biol (Mosk). 2007 May-Jun;41(3):433-49. Mol Biol (Mosk). 2007. PMID: 17685222 Review. Russian. - A structural perspective of CTD function.
Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. Meinhart A, et al. Genes Dev. 2005 Jun 15;19(12):1401-15. doi: 10.1101/gad.1318105. Genes Dev. 2005. PMID: 15964991 Review.
Cited by
- Repression of pervasive antisense transcription is the primary role of fission yeast RNA polymerase II CTD serine 2 phosphorylation.
Boulanger C, Haidara N, Yague-Sanz C, Larochelle M, Jacques PÉ, Hermand D, Bachand F. Boulanger C, et al. Nucleic Acids Res. 2024 Jul 22;52(13):7572-7589. doi: 10.1093/nar/gkae436. Nucleic Acids Res. 2024. PMID: 38801067 Free PMC article. - The RNA polymerase II CTD coordinates transcription and RNA processing.
Hsin JP, Manley JL. Hsin JP, et al. Genes Dev. 2012 Oct 1;26(19):2119-37. doi: 10.1101/gad.200303.112. Genes Dev. 2012. PMID: 23028141 Free PMC article. Review. - A genetic screen for terminator function in yeast identifies a role for a new functional domain in termination factor Nab3.
Loya TJ, O'Rourke TW, Reines D. Loya TJ, et al. Nucleic Acids Res. 2012 Aug;40(15):7476-91. doi: 10.1093/nar/gks377. Epub 2012 May 7. Nucleic Acids Res. 2012. PMID: 22564898 Free PMC article. - Reconstitution of mammalian cleavage factor II involved in 3' processing of mRNA precursors.
Schäfer P, Tüting C, Schönemann L, Kühn U, Treiber T, Treiber N, Ihling C, Graber A, Keller W, Meister G, Sinz A, Wahle E. Schäfer P, et al. RNA. 2018 Dec;24(12):1721-1737. doi: 10.1261/rna.068056.118. Epub 2018 Aug 23. RNA. 2018. PMID: 30139799 Free PMC article. - Nuclear mRNA quality control in yeast is mediated by Nrd1 co-transcriptional recruitment, as revealed by the targeting of Rho-induced aberrant transcripts.
Honorine R, Mosrin-Huaman C, Hervouet-Coste N, Libri D, Rahmouni AR. Honorine R, et al. Nucleic Acids Res. 2011 Apr;39(7):2809-20. doi: 10.1093/nar/gkq1192. Epub 2010 Nov 26. Nucleic Acids Res. 2011. PMID: 21113025 Free PMC article.
References
- Alen C., Kent,N.A., Jones,H.S., O’Sullivan,J., Aranda,A. and Proudfoot,N.J. (2002) A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell, 10, 1441–1452. - PubMed
- Aranda A. and Proudfoot,N. (2001) Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell, 7, 1003–1011. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous