Latent ClpX-recognition signals ensure LexA destruction after DNA damage - PubMed (original) (raw)
Latent ClpX-recognition signals ensure LexA destruction after DNA damage
Saskia B Neher et al. Genes Dev. 2003.
Abstract
The DNA-damage response genes in bacteria are up-regulated when LexA repressor undergoes autocatalytic cleavage stimulated by activated RecA protein. Intact LexA is stable to intracellular degradation but its auto-cleavage fragments are degraded rapidly. Here, both fragments of LexA are shown to be substrates for the ClpXP protease. ClpXP recognizes these fragments using sequence motifs that flank the auto-cleavage site but are dormant in intact LexA. Furthermore, ClpXP degradation of the LexA-DNA-binding fragment is important to cell survival after DNA damage. These results demonstrate how one protein-processing event can activate latent protease recognition signals, triggering a cascade of protein turnover in response to environmental stress.
Figures
Figure 1
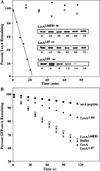
LexA auto-cleavage fragments are captured and degraded by ClpXP. (A) Western blot of the LexA species captured by the ClpPtrap and generated by auto-cleavage in vitro. (B) The proteolytic stability of LexA, LexA1–84, and LexA85–202 in vivo was determined in clpX+ (MC4100) and _clpX_− (SG22101) strains bearing plasmid pJL42. LexA, LexA1–84, and LexA85–202 were identified by comparison with auto-cleaved LexA (lane S). It is difficult to see the continued disappearance of LexA, as expected from auto-processing, because the Western blots are mildly overexposed to allow observation of the low levels of LexA85–202. (C) ClpXP degradation of purified LexA, LexA1–84, and LexA85–202 in vitro. The band marked ATP-RS is creatine kinase.
Figure 2
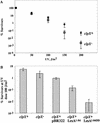
The new C terminus of LexA1–84 directs its degradation. (A) A time course of ClpXP degradation of LexA1–84, LexA1–87, and LexA1–84DD was quantified after SDS-PAGE. (B) ClpXP degradation of GFP-ssrA was measured by loss of fluorescence at 511 nm in the presence of various forms of LexA at 20 μM. An effective inhibitor of GFP-ssrA degradation, the ssrA peptide (20 μM), is included for comparison.
Figure 3
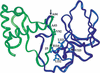
Location of latent ClpX-recognition signals in the structure of full-length LexA. The N-terminal domain is green and the C-terminal domain is blue. The proposed ClpX-recognition site in the C-terminal domain is labeled. The cleavage site is indicated by an arrow. (Structure from Luo et al. 2001.)
Figure 4
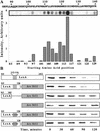
Identification of residues in LexA85–202 important for ClpXP recognition. (A) An array of LexA peptides was incubated with purified ClpX protein, and bound ClpX was detected using anti-ClpX antibody. The chart shows the relative intensity of ClpX binding to individual sequences. The starting amino acid positions of the 12-residue peptides are listed below the chart. The signal intensity of the peptide beginning at residue 115 may be artificially high. In other experiments, we have observed high signals from different peptides with the same three initial residues (VSG). (B) Degradation of LexA85–202 and LexA-Arc-st11 fusion proteins by ClpXP in vitro.
Figure 5
Cells accumulating LexA1–84 are UV sensitive. (A) Survival after UV irradiation of clpX+ (MC4100) and _clpX_− (SG22101) cells. The percentage of surviving cells is plotted against UV dose. Each point is the average (±S.D.) of three trials. (B) Survival of cells containing the empty parent vector or plasmids directing expression of LexA1–84 or LexA1–84DD was compared at a UV dose of 100 J/m2.
Similar articles
- Distinct peptide signals in the UmuD and UmuD' subunits of UmuD/D' mediate tethering and substrate processing by the ClpXP protease.
Neher SB, Sauer RT, Baker TA. Neher SB, et al. Proc Natl Acad Sci U S A. 2003 Nov 11;100(23):13219-24. doi: 10.1073/pnas.2235804100. Epub 2003 Oct 31. Proc Natl Acad Sci U S A. 2003. PMID: 14595014 Free PMC article. - The RssB response regulator directly targets sigma(S) for degradation by ClpXP.
Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. Zhou Y, et al. Genes Dev. 2001 Mar 1;15(5):627-37. doi: 10.1101/gad.864401. Genes Dev. 2001. PMID: 11238382 Free PMC article. - Cleavage of the Escherichia coli lexA protein by the recA protease.
Little JW, Edmiston SH, Pacelli LZ, Mount DW. Little JW, et al. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3225-9. doi: 10.1073/pnas.77.6.3225. Proc Natl Acad Sci U S A. 1980. PMID: 6447873 Free PMC article. - Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease.
Little JW. Little JW. Biochimie. 1991 Apr;73(4):411-21. doi: 10.1016/0300-9084(91)90108-d. Biochimie. 1991. PMID: 1911941 Review. - Pathways of protein remodeling by Escherichia coli molecular chaperones.
Pak M, Wickner SH. Pak M, et al. Genet Eng (N Y). 1996;18:203-17. doi: 10.1007/978-1-4899-1766-9_12. Genet Eng (N Y). 1996. PMID: 8785122 Review. No abstract available.
Cited by
- Staphylococcus aureus SOS response: Activation, impact, and drug targets.
Cheng K, Sun Y, Yu H, Hu Y, He Y, Shen Y. Cheng K, et al. mLife. 2024 Sep 30;3(3):343-366. doi: 10.1002/mlf2.12137. eCollection 2024 Sep. mLife. 2024. PMID: 39359682 Free PMC article. Review. - The LexA-RecA* structure reveals a cryptic lock-and-key mechanism for SOS activation.
Cory MB, Li A, Hurley CM, Carman PJ, Pumroy RA, Hostetler ZM, Perez RM, Venkatesh Y, Li X, Gupta K, Petersson EJ, Kohli RM. Cory MB, et al. Nat Struct Mol Biol. 2024 Oct;31(10):1522-1531. doi: 10.1038/s41594-024-01317-3. Epub 2024 May 16. Nat Struct Mol Biol. 2024. PMID: 38755298 - Diel Cycle Proteomics: Illuminating Molecular Dynamics in Purple Bacteria for Optimized Biotechnological Applications.
Matallana-Surget S, Geron A, Decroo C, Wattiez R. Matallana-Surget S, et al. Int J Mol Sci. 2024 Mar 2;25(5):2934. doi: 10.3390/ijms25052934. Int J Mol Sci. 2024. PMID: 38474181 Free PMC article. - Toxin-based screening of C-terminal tags in Escherichia coli reveals the exceptional potency of ssrA-like degrons.
Beardslee PC, Schmitz KR. Beardslee PC, et al. bioRxiv [Preprint]. 2024 Apr 12:2024.01.29.576913. doi: 10.1101/2024.01.29.576913. bioRxiv. 2024. PMID: 38352471 Free PMC article. Preprint. - Degron-controlled protein degradation in Escherichia coli: New Approaches and Parameters.
Cronan GE, Kuzminov A. Cronan GE, et al. bioRxiv [Preprint]. 2023 Nov 9:2023.11.08.566101. doi: 10.1101/2023.11.08.566101. bioRxiv. 2023. PMID: 37986802 Free PMC article. Updated. Preprint.
References
- Eguchi Y, Ogawa T, Ogawa H. Stimulation of RecA-mediated cleavage of phage phi 80 cI repressor by deoxydinucleotides. J Mol Biol. 1988;204:69–77. - PubMed
- Flynn, J.M., Neher, S.B., Kim, Y.I., Sauer, R.T., and Baker, T.A. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell (In press). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI016892/AI/NIAID NIH HHS/United States
- R01 GM049224/GM/NIGMS NIH HHS/United States
- R01 GM049224-11/GM/NIGMS NIH HHS/United States
- AI-16892/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources