Characterization of poly-gamma-glutamate hydrolase encoded by a bacteriophage genome: possible role in phage infection of Bacillus subtilis encapsulated with poly-gamma-glutamate - PubMed (original) (raw)
Characterization of poly-gamma-glutamate hydrolase encoded by a bacteriophage genome: possible role in phage infection of Bacillus subtilis encapsulated with poly-gamma-glutamate
Keitarou Kimura et al. Appl Environ Microbiol. 2003 May.
Abstract
Some Bacillus subtilis strains, including natto (fermented soybeans) starter strains, produce a capsular polypeptide of glutamate with a gamma-linkage, called poly-gamma-glutamate (gamma-PGA). We identified and purified a monomeric 25-kDa degradation enzyme for gamma-PGA (designated gamma-PGA hydrolase, PghP) from bacteriophage PhiNIT1 in B. subtilis host cells. The monomeric PghP internally hydrolyzed gamma-PGA to oligopeptides, which were then specifically converted to tri-, tetra-, and penta-gamma-glutamates. Monoiodoacetate and EDTA both inhibited the PghP activity, but Zn(2+) or Mn(2+) ions fully restored the enzyme activity inhibited by the chelator, suggesting that a cysteine residue(s) and these metal ions participate in the catalytic mechanism of the enzyme. The corresponding pghP gene was cloned and sequenced from the phage genome. The deduced PghP sequence (208 amino acids) with a calculated M(r) of 22,939 was not significantly similar to any known enzyme. Thus, PghP is a novel gamma-glutamyl hydrolase. Whereas phage PhiNIT1 proliferated in B. subtilis cells encapsulated with gamma-PGA, phage BS5 lacking PghP did not survive well on such cells. Moreover, all nine phages that contaminated natto during fermentation produced PghP, supporting the notion that PghP is important in the infection of natto starters that produce gamma-PGA. Analogous to polysaccharide capsules, gamma-PGA appears to serve as a physical barrier to phage absorption. Phages break down the gamma-PGA barrier via PghP so that phage progenies can easily establish infection in encapsulated cells.
Figures
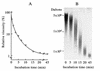
FIG. 1.
γ-PGA degradation activity in a B. subtilis NAFM5 culture infected with ΦNIT1 phage measured by viscometry (A) and by agarose gel electrophoresis (B). Reactions proceeded as described in Materials and Methods, and portions of mixtures were withdrawn after the indicated incubation periods. (A) Viscosity was measured using a falling-ball viscometer and is expressed as relative viscosity to the mixture at zero time. (B) Samples (10 μl) were resolved by 1.0% agarose gel electrophoresis, and degradation products were visualized by staining with methylene blue. Positions of intact γ-PGA (5 × 106 Da) and of 104 Da and 103 Da γ-PGA (fractionated from partial hydrolysates of γ-PGA by PghP through gel filtration and high-performance liquid chromatography) (21) are indicated at left.
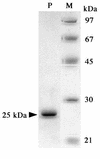
FIG. 2.
SDS-PAGE of purified PghP. PghP (1 μg, lane P) from Superose 12 column chromatography was analyzed together with molecular mass markers (1 μg each, lane M) by SDS-10% PAGE (17). Molecular markers: phosphorylase b (97 kDa), bovine serum albumin (67 kDa), ovalbumin (45 kDa), carbonic anhydrase (30 kDa), and catalase (21 kDa).
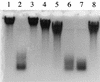
FIG. 3.
Effects of monoiodoacetate, EDTA, and divalent cations on PghP activity. Reactions proceeded at 37°C for 60 min in PghP assay mixture (see Materials and Methods) containing 50 mM Tris-HCl (pH 7.5) instead of 50 mM sodium phosphate (pH 7.5), and reaction products were analyzed using agarose gel electrophoresis. Lane 1, no enzyme (control). Lanes 2 through 8 contained 45 ng of purified PghP/ml plus the following: lane 2, no further addition; lane 3, 1 mM EDTA; lane 4, 1 mM EDTA + 5 mM CaCl2; lane 5, 1 mM EDTA + 5 mM MgCl2; lane 6, 1 mM EDTA + 5 mM MnCl2, lane 7, 1 mM EDTA + 5 mM ZnCl2; lane 8, 1 mM monoiodoacetate. Reaction mixtures for lanes 4 to 7 were incubated for 5 min before adding chlorides.
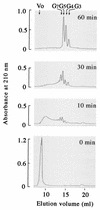
FIG. 4.
Analysis of γ-PGA degradation products generated by PghP over reaction time. Reactions proceeded under standard assay conditions (see Materials and Methods). Samples (0.2 ml) after various incubation periods were applied onto Superdex peptide columns, and reaction products were monitored by absorbance at 210 nm. Molecular sizes of oligomers were estimated from elution volumes relative to those of glutamate (_M_r = 147), di-γ-glutamate (_M_r = 276), tetra-γ-glutamate (_M_r = 535), and gastrin (_M_r = 2,126). Abbreviations: Vo, void volume; G3, tri-γ-glutamate; G4, tetra-γ-glutamate; G5, penta-γ-glutamate; G7, hepta-γ-glutamate.
FIG. 5.
Location of pghP on an _Eco_RV fragment and the amino-terminal region of PghP. Deduced and determined amino-terminal sequences match perfectly (underline). Possible ribosome-binding site (GGAGG) precedes initiation ATG codon at 9 bp upstream. Nucleotides 5′-TA(C/T)CCGAA(C/T)ATTGA(A/G)GC-3′ used in Southern blotting and in cloning pghP correspond to region between residues 7 and 12 (boxed) of deduced amino-terminal sequence.
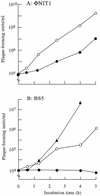
FIG. 6.
Effects of γ-PGA capsule on growth of phages with or without PghP. B. subtilis NAFM5 was shaken in GSP medium (22) at 37°C. When the optical density at 600 nm of the cultures reached 0.3 (noncapsulated [open circles]) or after 24 h (encapsulated stationary phase [closed circles]), phages ΦNIT1 (PghP+) (A) and BS5 (PghP−) (B) were added at concentrations of 104 PFU/ml and incubation was continued at 37°C. Purified PghP (1 μg/ml) was added to stationary-phase cultures together with BS5 (closed triangle). After indicated periods, phages in cultures were titrated against strain NAFM5 as the indicator.
Similar articles
- γ-PGA Hydrolases of Phage Origin in Bacillus subtilis and Other Microbial Genomes.
Mamberti S, Prati P, Cremaschi P, Seppi C, Morelli CF, Galizzi A, Fabbi M, Calvio C. Mamberti S, et al. PLoS One. 2015 Jul 9;10(7):e0130810. doi: 10.1371/journal.pone.0130810. eCollection 2015. PLoS One. 2015. PMID: 26158264 Free PMC article. - Genomic analysis of Bacillus subtilis lytic bacteriophage ϕNIT1 capable of obstructing natto fermentation carrying genes for the capsule-lytic soluble enzymes poly-γ-glutamate hydrolase and levanase.
Ozaki T, Abe N, Kimura K, Suzuki A, Kaneko J. Ozaki T, et al. Biosci Biotechnol Biochem. 2017 Jan;81(1):135-146. doi: 10.1080/09168451.2016.1232153. Epub 2016 Sep 22. Biosci Biotechnol Biochem. 2017. PMID: 27885938 - Complete nucleotide sequence of Bacillus subtilis (natto) bacteriophage PM1, a phage associated with disruption of food production.
Umene K, Shiraishi A. Umene K, et al. Virus Genes. 2013 Jun;46(3):524-34. doi: 10.1007/s11262-013-0876-4. Epub 2013 Jan 13. Virus Genes. 2013. PMID: 23315235 - Biochemistry and molecular genetics of poly-gamma-glutamate synthesis.
Ashiuchi M, Misono H. Ashiuchi M, et al. Appl Microbiol Biotechnol. 2002 Jun;59(1):9-14. doi: 10.1007/s00253-002-0984-x. Epub 2002 Apr 16. Appl Microbiol Biotechnol. 2002. PMID: 12073126 Review. - DNA bending and looping in the transcriptional control of bacteriophage phi29.
Camacho A, Salas M. Camacho A, et al. FEMS Microbiol Rev. 2010 Sep;34(5):828-41. doi: 10.1111/j.1574-6976.2010.00219.x. Epub 2010 Mar 17. FEMS Microbiol Rev. 2010. PMID: 20412311 Review.
Cited by
- Genomic diversity and comprehensive taxonomical classification of 61 Bacillus subtilis group member infecting bacteriophages, and the identification of ortholog taxonomic signature genes.
Abraha HB, Lee JW, Kim G, Ferdiansyah MK, Ramesha RM, Kim KP. Abraha HB, et al. BMC Genomics. 2022 Dec 16;23(1):835. doi: 10.1186/s12864-022-09055-w. BMC Genomics. 2022. PMID: 36526963 Free PMC article. - The structure of PghL hydrolase bound to its substrate poly-γ-glutamate.
Ramaswamy S, Rasheed M, Morelli CF, Calvio C, Sutton BJ, Pastore A. Ramaswamy S, et al. FEBS J. 2018 Dec;285(24):4575-4589. doi: 10.1111/febs.14688. Epub 2018 Nov 19. FEBS J. 2018. PMID: 30387270 Free PMC article. - Isolation and Characterization of Phages Infecting Bacillus subtilis.
Krasowska A, Biegalska A, Augustyniak D, Łoś M, Richert M, Łukaszewicz M. Krasowska A, et al. Biomed Res Int. 2015;2015:179597. doi: 10.1155/2015/179597. Epub 2015 Jul 26. Biomed Res Int. 2015. PMID: 26273592 Free PMC article. - Poly-gamma-glutamate capsule-degrading enzyme treatment enhances phagocytosis and killing of encapsulated Bacillus anthracis.
Scorpio A, Chabot DJ, Day WA, O'brien DK, Vietri NJ, Itoh Y, Mohamadzadeh M, Friedlander AM. Scorpio A, et al. Antimicrob Agents Chemother. 2007 Jan;51(1):215-22. doi: 10.1128/AAC.00706-06. Epub 2006 Oct 30. Antimicrob Agents Chemother. 2007. PMID: 17074794 Free PMC article. - Genomic analysis of Pseudomonas putida phage tf with localized single-strand DNA interruptions.
Glukhov AS, Krutilina AI, Shlyapnikov MG, Severinov K, Lavysh D, Kochetkov VV, McGrath JW, de Leeuwe C, Shaburova OV, Krylov VN, Akulenko NV, Kulakov LA. Glukhov AS, et al. PLoS One. 2012;7(12):e51163. doi: 10.1371/journal.pone.0051163. Epub 2012 Dec 7. PLoS One. 2012. PMID: 23236447 Free PMC article.
References
- Ackermann, H.-W., R. R. Azizbekyan, R. L. Bernier, H. de Barje, S. Saindouk, J.-R. Valéro, and M.-X. Yu. 1995. Phage typing of Bacillus subtilis and B. thuringensis. Res. Microbiol. 146:643-657. - PubMed
- Ashiuchi, M., C. Nawa, T. Kamei, J.-J. Song, S.-P. Hong, M.-H. Sung, K. Soda, T. Yagi, and H. Misono. 2001. Physical and biochemical characterization of poly-γ-glutamate synthase complex of Bacillus subtilis. Eur. J. Biochem. 268:5321-5328. - PubMed
- Ashiuchi, M., K. Soda, and H. Misono. 1999. A poly-γ-glutamate synthetic complex of Bacillus subtilis IFO3336: gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 263:6-12. - PubMed
- Bayer, M. E., H. Thurow, and M. H. Bayer. 1979. Penetration of the polysaccharide capsule of Escherichia coli (Bi161/42) by bacteriophage K29. Virology 94:95-118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources