The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation - PubMed (original) (raw)
The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation
Daniel Schaft et al. Nucleic Acids Res. 2003.
Abstract
Existing evidence indicates that SET2, the histone 3 lysine 36 methyltransferase of Saccharomyces cerevisiae, is a transcriptional repressor. Here we show by five main lines of evidence that SET2 is involved in transcriptional elongation. First, most, if not all, subunits of the RNAP II holoenzyme co-purify with SET2. Second, all of the co-purifying RNAP II subunit, RPO21, was phosphorylated at serines 5 and 2 of the C-terminal domain (CTD) tail, indicating that the SET2 association is specific to either the elongating or SSN3 repressed forms (or both) of RNAP II. Third, the association of SET2 with CTD phosphorylated RPO21 remained in the absence of ssn3. Fourth, in the absence of ssn3, mRNA production from gal1 required SET2. Fifth, SET2 was detected on gal1 by in vivo crosslinking after, but not before, the induction of transcription. Similarly, SET2 physically associated with the transcribed region of pdr5 but was not detected on gal1 or pdr5 promoter regions. Since SET2 is also a histone methyltransferase, these results suggest a role for histone 3 lysine 36 methylation in transcriptional elongation.
Figures
Figure 1
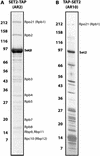
TAP purification of SET2. Affinity purified SET2 was separated on 7–25% SDS–PAGE and visualized by staining with Coomassie blue. Molecular weight markers indicated on the left are in kilodaltons. All bands present in these gels were identified and only the specific ones are labeled. (A) The TAP tag was fused to the C-terminus of SET2 (SET2-TAP). (B) The TAP tag was fused to the N-terminus of SET2 (TAP-SET2).
Figure 2
Biochemical properties of cellular SET2-TAP. Total cell extracts were fractionated on a Superose 6 sizing column before analysis by western blotting using a secondary antibody (PAP) to the TAP tag or antibodies specific for RPO21 of RNAP II, as indicated. (A) Elution profiles of SET2-TAP and RNAP II detected by PAP (SET2-TAP) or antibodies specific for unphosphorylated (8WG16) and phosphorylated (CTD4H8) RPO21. Molecular weights, assuming globular Stokes radii, were estimated from parallel size standards. (B) Estimation of cellular abundance of SET2-TAP by dot blotting. A standard curve was made using highly purified GST–TAP that had been quantitated by Bradford. Either 80 or 8 ng were spotted followed by a three-fold dilution series as indicated. Ten milligrams of total yeast proteins isolated from wild-type or SET2-TAP strains were spotted at the left as indicated, followed by three-fold dilutions. (C) Graph of the standard curve. The two highest points (80 and 27 ng) fall below the line due to saturation in the assay and have been omitted. Given 200 pg total protein per cell, we estimate 35 000 SET2 molecules/cell.
Figure 3
SET2 associates with the elongating, CTD phosphorylated, RPO21. (A) Total cell extracts from the N-terminal tagged TAP-SET2 or SIF2-TAP strains were either directly used (input) or affinity purified (purified) before electrophoresis and blotting. The blots were probed with antibodies detecting unphosphorylated (8WG16, Covance) and serine 5 phosphorylated (CTD4H8, Upstate) RPO21. Ponceau staining of the same blots showing the RPO21 bands on both membranes. (B) As for (A) except that C-terminal tagged SET2 was used from either a wild-type background or a Δssn3 background as indicated. Also, parallel blots were probed with antibodies against RPO21, whether phosphorylated or not, (ARNA3a, Research Diagnostics), serine 2 phosphorylated (H14, Covance) and serine 5 phosphorylated (H5, Covance) RPO21.
Figure 4
Sensitivities of strains carrying deletions of set2 and ssn3 to heat and 6-AU. Yeast strains carrying deletions of set2, ssn3 or neither were spotted onto minimal media plates with or without 6-AU and cultured at three temperatures, as indicated.
Figure 5
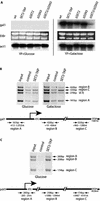
SET2 is a transcriptional elongation factor. (A) Northern blots of total RNA prepared from wt, SET2-TAP, Δset2, Δssn3 or Δset2/Δssn3 strains, grown in glucose- or galactose-containing media as indicated, were hybridized with probes from the gal1 or act1 genes. In the center, the ethidium bromide stained gel for the gal1 blot is shown. (B) ChIPs were performed with extracts, prepared according to Hecht and Grunstein (20), using rabbit IgG agarose beads to immunoprecipitate SET2-TAP or empty agarose beads (control). Input represents signals from the total extract without immunoprecipitation. Three primer pairs to amplify regions of the gal1 gene are depicted below on a diagram of the gene, with the sizes of the amplification products indicated. The positions of the amplified regions with respect to the position of the initiating methionine are also indicated. Primer pairs for a telomeric region of chromosome VI (VI R) were included. (C) As for (B) except that the pdr5 gene was investigated.
Similar articles
- Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation.
Li J, Moazed D, Gygi SP. Li J, et al. J Biol Chem. 2002 Dec 20;277(51):49383-8. doi: 10.1074/jbc.M209294200. Epub 2002 Oct 14. J Biol Chem. 2002. PMID: 12381723 - A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation.
Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. Kizer KO, et al. Mol Cell Biol. 2005 Apr;25(8):3305-16. doi: 10.1128/MCB.25.8.3305-3316.2005. Mol Cell Biol. 2005. PMID: 15798214 Free PMC article. - Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II.
Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Krogan NJ, et al. Mol Cell Biol. 2003 Jun;23(12):4207-18. doi: 10.1128/MCB.23.12.4207-4218.2003. Mol Cell Biol. 2003. PMID: 12773564 Free PMC article. - Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation.
Hampsey M, Reinberg D. Hampsey M, et al. Cell. 2003 May 16;113(4):429-32. doi: 10.1016/s0092-8674(03)00360-x. Cell. 2003. PMID: 12757703 Review. - A site to remember: H3K36 methylation a mark for histone deacetylation.
Lee JS, Shilatifard A. Lee JS, et al. Mutat Res. 2007 May 1;618(1-2):130-4. doi: 10.1016/j.mrfmmm.2006.08.014. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17346757 Review.
Cited by
- Mechanism of transcription through a nucleosome by RNA polymerase II.
Kulaeva OI, Hsieh FK, Chang HW, Luse DS, Studitsky VM. Kulaeva OI, et al. Biochim Biophys Acta. 2013 Jan;1829(1):76-83. doi: 10.1016/j.bbagrm.2012.08.015. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982194 Free PMC article. Review. - Systematic perturbations of SETD2, NSD1, NSD2, NSD3, and ASH1L reveal their distinct contributions to H3K36 methylation.
Shipman GA, Padilla R, Horth C, Hu B, Bareke E, Vitorino FN, Gongora JM, Garcia BA, Lu C, Majewski J. Shipman GA, et al. Genome Biol. 2024 Oct 10;25(1):263. doi: 10.1186/s13059-024-03415-3. Genome Biol. 2024. PMID: 39390582 Free PMC article. - reSETting chromatin during transcription elongation.
Smolle M, Workman JL, Venkatesh S. Smolle M, et al. Epigenetics. 2013 Jan;8(1):10-5. doi: 10.4161/epi.23333. Epub 2012 Dec 20. Epigenetics. 2013. PMID: 23257840 Free PMC article. Review. - Mediator subunits and histone methyltransferase Set2 contribute to Ino2-dependent transcriptional activation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae.
Dettmann A, Jäschke Y, Triebel I, Bogs J, Schröder I, Schüller HJ. Dettmann A, et al. Mol Genet Genomics. 2010 Mar;283(3):211-21. doi: 10.1007/s00438-009-0508-9. Mol Genet Genomics. 2010. PMID: 20054697 - A combinatorial view of old and new RNA polymerase II modifications.
Lyons DE, McMahon S, Ott M. Lyons DE, et al. Transcription. 2020 Apr;11(2):66-82. doi: 10.1080/21541264.2020.1762468. Epub 2020 May 13. Transcription. 2020. PMID: 32401151 Free PMC article. Review.
References
- Lachner M. and Jenuwein,T. (2002) The many faces of histone lysine methylation. Curr. Opin. Cell Biol., 14, 286–298. - PubMed
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. - PubMed
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
- Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. - PubMed
- Ringrose L. and Paro,R. (2001) Gene regulation. Cycling silence. Nature, 412, 493–494. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials