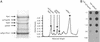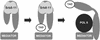TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution - PubMed (original) (raw)
TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution
Camilla O Samuelsen et al. Proc Natl Acad Sci U S A. 2003.
Abstract
In Saccharomyces cerevisiae Mediator, a subgroup of proteins (Srb8, Srb9, Srb10, and Srb11) form a module, which is involved in negative regulation of transcription. Homologues of Srb10 and Srb11 are found in some mammalian Mediator preparations, whereas no clear homologues have been reported for Srb8 and Srb9. Here, we identify a TRAP240/ARC250 homologue in Schizosaccharomyces pombe and demonstrate that this protein, spTrap240, is stably associated with a larger form of Mediator, which also contains conserved homologues of Srb8, Srb10, and Srb11. We find that spTrap240 and Sch. pombe Srb8 (spSrb8) regulate the same distinct subset of genes and have indistinguishable phenotypic characteristics. Importantly, Mediator containing the spSrb8/spTrap240/spSrb10/spSrb11 subunits is isolated only in free form, devoid of RNA polymerase II. In contrast, Mediator lacking this module associates with the polymerase. Our findings provide experimental evidence for recent suggestions that TRAP230/ARC240 and TRAP240/ARC250 may indeed be the Srb8 and Srb9 homologues of mammalian Mediator. Apparently Srb8/TRAP230/ARC240, Srb9/TRAP240/ARC250, Srb10, and Srb11 constitute a conserved Mediator submodule, which is involved in negative regulation of transcription in all eukaryotes.
Figures
Fig. 1.
Substoichiometric amounts of spSrb8 and spTrap240 in Mediator prepared from Sch. pombe.(A left) High molecular weight region of Mediator purified over IgG, from TAP-spMed7 cells. Proteins were separated by SDS/10% PAGE and revealed by staining with Coomassie blue. (Right) Densitometry of the SDS gel showed that the Mediator preparation contained substoichiometric amounts of Rbp1, Rbp2, spSrb8, and spTrap240 compared with the core Mediator subunit spRgr1/Pmc1. (B) Dot blotting with rabbit anti-peroxidase IgG of dilution series of whole-cell extracts from Sch. pombe (TAP-spMed7 and TAP-spTrap240). Crude extracts were prepared and blotted as described. The protein quality in our extracts was verified by separation of proteins on an SDS/10% polyacrylamide gel and immunoblot analysis.
Fig. 2.
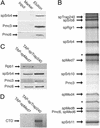
A spTrap240 containing Mediator complex. (A) TAP-Trap240 copurifies with Mediator proteins over IgG Sepharose. We analyzed the input (whole-cell extract, 15 μl) and the eluted proteins from the IgG Sepharose (15 μl) by immunoblotting. (B) After TAP purification, the TAP-spTrap240-containing Mediator complex was analyzed in an SDS/10% polyacrylamide gel and revealed by silver staining. The two bands denoted with asterisks (*) are contaminants corresponding to ribosomal proteins. (C) Immunoblotting reveals that Rpb1 is present in TAP-spMed7-purified Mediator, but is absent in the TAP-spTrap240 Mediator preparation. (D) CTD kinase assay with TAP-spMed7 and TAP-spTrap240 Mediator preparations. Purified S. cerevisiae pol II (100 ng per reaction) was used as substrate.
Fig. 3.
Alignment of spTrap240 and homologues from higher eukaryotic cells. The database accession codes for the proteins are NP_594050 (Sch. pombe), NP_005112 (Homo sapiens), and AAF50591 (D. melanogaster). The Srb9 sequence from C. albicans was obtained by an analysis of unfinished sequence data from the C. albicans genome project (
www-sequence.stanford.edu/group/candida/index.html
).
Fig. 4.
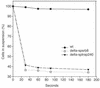
Flocculation level of Δ_spsrb8_, Δ_sptrap240_, and WT Sch. pombe under rich growth conditions. The cell number in the supernatant was determined by an OD at 600 nm at the indicated times. Both Δ_spsrb8_ and Δ_sptrap240_ are highly flocculent.
Fig. 5.
A transcription activation domain (TAD) interacts directly with Srb8–11-containing Mediator and recruits the complex to specific promoters. The Srb8–11 complex is degraded at the promoter, allowing interactions between Mediator and pol II.
Similar articles
- A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast.
Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD. Borggrefe T, et al. J Biol Chem. 2002 Nov 15;277(46):44202-7. doi: 10.1074/jbc.M207195200. Epub 2002 Aug 27. J Biol Chem. 2002. PMID: 12200444 - The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription.
Larschan E, Winston F. Larschan E, et al. Mol Cell Biol. 2005 Jan;25(1):114-23. doi: 10.1128/MCB.25.1.114-123.2005. Mol Cell Biol. 2005. PMID: 15601835 Free PMC article. - Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions.
Zhu X, Wirén M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Zhu X, et al. Mol Cell. 2006 Apr 21;22(2):169-78. doi: 10.1016/j.molcel.2006.03.032. Mol Cell. 2006. PMID: 16630887 - Mediator influences Schizosaccharomyces pombe RNA polymerase II-dependent transcription in vitro.
Spahr H, Khorosjutina O, Baraznenok V, Linder T, Samuelsen CO, Hermand D, Mäkela TP, Holmberg S, Gustafsson CM. Spahr H, et al. J Biol Chem. 2003 Dec 19;278(51):51301-6. doi: 10.1074/jbc.M306750200. Epub 2003 Oct 8. J Biol Chem. 2003. PMID: 14534314 - Yeast 14-3-3 proteins.
van Heusden GP, Steensma HY. van Heusden GP, et al. Yeast. 2006 Feb;23(3):159-71. doi: 10.1002/yea.1338. Yeast. 2006. PMID: 16498703 Review.
Cited by
- Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo.
Jeronimo C, Langelier MF, Bataille AR, Pascal JM, Pugh BF, Robert F. Jeronimo C, et al. Mol Cell. 2016 Nov 3;64(3):455-466. doi: 10.1016/j.molcel.2016.09.002. Epub 2016 Oct 20. Mol Cell. 2016. PMID: 27773677 Free PMC article. - Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs.
Guo Z, Stiller JW. Guo Z, et al. BMC Genomics. 2004 Sep 20;5:69. doi: 10.1186/1471-2164-5-69. BMC Genomics. 2004. PMID: 15380029 Free PMC article. - Cyclin-dependent kinases: Masters of the eukaryotic universe.
Pluta AJ, Studniarek C, Murphy S, Norbury CJ. Pluta AJ, et al. Wiley Interdiscip Rev RNA. 2023 Sep 17;15(1):e1816. doi: 10.1002/wrna.1816. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37718413 Free PMC article. Review. - Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants.
Samanta S, Thakur JK. Samanta S, et al. Front Plant Sci. 2015 Sep 17;6:757. doi: 10.3389/fpls.2015.00757. eCollection 2015. Front Plant Sci. 2015. PMID: 26442070 Free PMC article. Review. - Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Li G, Snyder L, Nakanishi K, Chang CE, Murakami K, Garcia BA, Boyer TG, Tsai KL. Li YC, et al. Sci Adv. 2021 Jan 15;7(3):eabd4484. doi: 10.1126/sciadv.abd4484. Print 2021 Jan. Sci Adv. 2021. PMID: 33523904 Free PMC article.
References
- Kim, Y. J., Bjorklund, S., Li, Y., Sayre, M. H. & Kornberg, R. D. (1994) Cell 77, 599–608. - PubMed
- Myers, L. C. & Kornberg, R. D. (2000) Annu. Rev. Biochem. 69, 729–749. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous