PPARgamma and PPARdelta negatively regulate specific subsets of lipopolysaccharide and IFN-gamma target genes in macrophages - PubMed (original) (raw)
PPARgamma and PPARdelta negatively regulate specific subsets of lipopolysaccharide and IFN-gamma target genes in macrophages
John S Welch et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Natural and synthetic agonists of the peroxisome proliferator-activated receptor gamma (PPARgamma) regulate adipocyte differentiation, glucose homeostasis, and inflammatory responses. Although effects on adipogenesis and glucose metabolism are genetically linked to PPARgamma, the PPARgamma dependence of antiinflammatory responses of these substances is less clear. Here, we have used a combination of mRNA expression profiling and conditional disruption of the PPARgamma gene in mice to characterize programs of transcriptional activation and repression by PPARgamma agonists in elicited peritoneal macrophages. Natural and synthetic PPARgamma agonists, including the thiazolidinedione rosiglitazone (Ro), modestly induced the expression of a surprisingly small number of genes, several of which were also induced by a specific PPARdelta agonist. The majority of these genes encode proteins involved in lipid homeostasis. In contrast, Ro inhibited induction of broad subsets of lipopolysaccharide and IFN-gamma target genes in a gene-specific and PPARgamma-dependent manner. At high concentrations, Ro inhibited induction of lipopolysaccharide target genes in PPARgamma-deficient macrophages, at least in part by activating PPARdelta. These studies establish overlapping transactivation and transrepression functions of PPARgamma and PPARdelta in macrophages and suggest that a major transcriptional role of PPARgamma is negative regulation of specific subsets of genes that are activated by T helper 1 cytokines and pathogenic molecules that signal through pattern recognition receptors. These findings support a physiological role of PPARgamma in regulating both native and acquired immune responses.
Figures
Fig. 1.
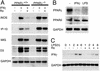
Ro modestly induces a small set of genes in thioglycollate-elicited macrophages. (A) Scatter plot of mRNA expression levels as assessed by hybridization of cRNA from macrophages treated for 24 h with Ro (10 μM) or control solvent. Genes confirmed to be induced by secondary analysis are indicated by colored data points. (B) RT-PCR analysis of PPARγ mRNA from peritoneal macrophages of PPARγf/f and Mx-Cre+/PPARγf/f mice treated with polyinosinic-polycytidylic acid (pIpC) illustrates quantitative excision of exon 2. (C) Dendogram of genes represented on the Affymetrix U74A microarray found to be reproducibly induced by Ro in wild-type peritoneal macrophages. Red indicates up-regulation and green indicates down-regulation with respect to levels of expression in untreated PPARγ+/+ macrophages. The colors of gene names in C correspond to the colors of data points in A.
Fig. 2.
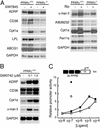
PPARγ and PPARδ positively regulate an overlapping set of genes involved in lipid metabolism. (A) Secondary analysis of positively regulated genes by Northern blotting. PPARγ+/+ or PPARγ–/– macrophages were treated for 24 h with control solvent or 1 μM concentrations of either Ro or the PPARγ-specific agonist GW7845. RNA was harvested and 10 μg was analyzed by Northern blotting with specific probes for the indicated mRNAs. (B) PPARδ activates an overlapping set of genes. PPARγ+/+ macrophages were treated for 24 h with control solvent, or 0.1 μMor1 μM of the PPARδ-specific agonist GW0742. RNA was analyzed by Northern blotting as described above. (C) Ro can induce gene expression through PPARδ. RAW 264.7 cells that lack PPARγ were transfected with a PPAR-responsive reporter gene. Cells were cotransfected with CMV expression vectors and treated with either GW0742 or Ro as follows: •, GW0742 + CMV-PPARδ; ○, GW0742 + CMV vector; ▪, Ro + CMV-PPARδ; □, Ro + CMV vector. Luciferase activity was assayed 36 h after drug treatment.
Fig. 3.
Inhibition of LPS-dependent gene expression by Ro. (A) Dendogram of genes in which LPS induction was inhibited >40% by Ro. The color scheme for increased or decreased expression is the same as described in Fig. 1. (B) Confirmation of negative regulation of LPS target genes by Northern blotting.
Fig. 4.
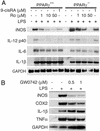
PPARγ and PPARδ mediate inhibitory effects of Ro. (A) Inhibition of LPS-dependent expression of iNOS and IL-12 p40 by Ro is PPARγ dependent at 1 μM and PPARγ independent at 50 μM. PPARγ+/+ and PPARγ–/– macrophages were treated with LPS (100 ng/ml) for 6 h in the presence of the indicated concentrations of Ro. Ten micrograms of total RNA was analyzed for expression of the indicated genes by Northern blotting. (B) PPARδ potently inhibits LPS-dependent induction of iNOS and COX2. PPARγ+/+ macrophages were treated with LPS for6hinthe presence of the indicated concentrations of the PPARδ agonist GW0742. Ten micrograms of total RNA was analyzed for expression of the indicated genes by Northern blotting.
Fig. 5.
Mutual antagonism of IFN-γ and PPARγ signaling pathways. (A) Ro inhibits IFN-γ induction of iNOS in a PPARγ-dependent manner. PPARγ+/+ and PPARγ–/– macrophages were treated with IFN-γ (100 units/ml) for 6 h in the presence of Ro (10 μM). (B) PPARγ expression but not PPARδ expression is inhibited in peritoneal macrophages by IFN-γ and LPS. PPARγ+/+ macrophages were treated for 6 h with either IFN-γ or LPS before analysis of mRNA. (C) Ro inhibits induction of IFN-γ expression in macrophages by LPS. Cells were treated for the indicated times with LPS in the presence or absence of Ro (10 μM). In all experiments depicted here, 10 μg of total RNA was analyzed for expression of the indicated genes by Northern blotting.
Comment in
- Suppression of macrophage inflammatory responses by PPARs.
Henson P. Henson P. Proc Natl Acad Sci U S A. 2003 May 27;100(11):6295-6. doi: 10.1073/pnas.1232410100. Epub 2003 May 19. Proc Natl Acad Sci U S A. 2003. PMID: 12756292 Free PMC article. No abstract available.
Similar articles
- Pro-inflammatory cytokines negatively regulate PPARγ mediated gene expression in both human and murine macrophages via multiple mechanisms.
Nagy ZS, Czimmerer Z, Szanto A, Nagy L. Nagy ZS, et al. Immunobiology. 2013 Nov;218(11):1336-44. doi: 10.1016/j.imbio.2013.06.011. Epub 2013 Jul 1. Immunobiology. 2013. PMID: 23870825 - The peroxisome proliferator-activated receptor beta/delta agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells.
Dressel U, Allen TL, Pippal JB, Rohde PR, Lau P, Muscat GE. Dressel U, et al. Mol Endocrinol. 2003 Dec;17(12):2477-93. doi: 10.1210/me.2003-0151. Epub 2003 Oct 2. Mol Endocrinol. 2003. PMID: 14525954 - The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis.
Hummasti S, Tontonoz P. Hummasti S, et al. Mol Endocrinol. 2006 Jun;20(6):1261-75. doi: 10.1210/me.2006-0025. Epub 2006 Mar 23. Mol Endocrinol. 2006. PMID: 16556736 - Peroxisome proliferator-activated receptor-gamma in macrophage lipid homeostasis.
Lee CH, Evans RM. Lee CH, et al. Trends Endocrinol Metab. 2002 Oct;13(8):331-5. doi: 10.1016/s1043-2760(02)00668-9. Trends Endocrinol Metab. 2002. PMID: 12217489 Review. - Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation.
Chinetti G, Fruchart JC, Staels B. Chinetti G, et al. Inflamm Res. 2000 Oct;49(10):497-505. doi: 10.1007/s000110050622. Inflamm Res. 2000. PMID: 11089900 Review.
Cited by
- PPARs and molecular mechanisms of transrepression.
Ricote M, Glass CK. Ricote M, et al. Biochim Biophys Acta. 2007 Aug;1771(8):926-35. doi: 10.1016/j.bbalip.2007.02.013. Epub 2007 Mar 12. Biochim Biophys Acta. 2007. PMID: 17433773 Free PMC article. Review. - Role of JAK-STAT and PPAR-Gamma Signalling Modulators in the Prevention of Autism and Neurological Dysfunctions.
Khera R, Mehan S, Kumar S, Sethi P, Bhalla S, Prajapati A. Khera R, et al. Mol Neurobiol. 2022 Jun;59(6):3888-3912. doi: 10.1007/s12035-022-02819-1. Epub 2022 Apr 18. Mol Neurobiol. 2022. PMID: 35437700 Review. - STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells.
Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, Barak Y, Schwabe J, Nagy L. Szanto A, et al. Immunity. 2010 Nov 24;33(5):699-712. doi: 10.1016/j.immuni.2010.11.009. Immunity. 2010. PMID: 21093321 Free PMC article. - Deletion of PPARγ in Mesenchymal Lineage Cells Protects Against Aging-Induced Cortical Bone Loss in Mice.
Cao J, Ding K, Pan G, Rosario R, Su Y, Bao Y, Zhou H, Xu J, McGee Lawrence ME, Hamrick MW, Isales CM, Shi X. Cao J, et al. J Gerontol A Biol Sci Med Sci. 2020 Apr 17;75(5):826-834. doi: 10.1093/gerona/glaa049. J Gerontol A Biol Sci Med Sci. 2020. PMID: 32060555 Free PMC article. - Electrophile Modulation of Inflammation: A Two-Hit Approach.
O'Brien J, Wendell SG. O'Brien J, et al. Metabolites. 2020 Nov 10;10(11):453. doi: 10.3390/metabo10110453. Metabolites. 2020. PMID: 33182676 Free PMC article. Review.
References
- Willson, T. M., Lambert, M. H. & Kliewer, S. A. (2001) Annu. Rev. Biochem. 70, 341–367. - PubMed
- Chawla, A., Repa, J., Evans, R. & Mangelsdorf, D. (2001) Science 294, 1866–1870. - PubMed
- Spiegelman, B. M. (1998) Diabetes 47, 507–514. - PubMed
- Xu, H. E., Lambert, M. H., Montana, V. G., Parks, D. J., Blanchard, S. G., Brown, P. J., Sternbach, D. D., Lehmann, J. M., Wisely, G. B., Willson, T. M., et al. (1999) Mol. Cell 3, 397–403. - PubMed
- Forman, B. M., Tontonoz, P., Chen, J., Brun, R. P., Spiegelman, B. M. & Evans, R. M. (1995) Cell 83, 803–812. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials