Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway - PubMed (original) (raw)
Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway
Antonio A Postigo. EMBO J. 2003.
Abstract
Binding of TGFbeta/BMP factors to their receptors leads to translocation of Smad proteins to the nucleus where they activate transcription of target genes. The two-handed zinc finger proteins encoded by Zfhx1a and Zfhx1b, ZEB-1/deltaEF1 and ZEB-2/SIP1, respectively, regulate gene expression and differentiation programs in a number of tissues. Here I demonstrate that ZEB proteins are also crucial regulators of TGFbeta/BMP signaling with opposing effects on this pathway. Both ZEB proteins bind to Smads, but while ZEB-1/deltaEF1 synergizes with Smad proteins to activate transcription, promote osteoblastic differentiation and induce cell growth arrest, the highly related ZEB-2/SIP1 protein has the opposite effect. Finally, the ability of TGFbeta to mediate transcription of TGFbeta-dependent genes and induce growth arrest depends on the presence of endogenous ZEB-1/deltaEF1 protein.
Figures
Fig. 1. (A) Schematic representation of the ZEB family (ZEB-1/δEF1 and ZEB-2/SIP1) of two-handed zinc finger factors. The central region (CR) in between both zinc finger clusters act as a repressor domain in part through the recruitment of the CtBP corepressor through a CID. 3′ of the N-terminal zinc fingers there is a region that acts as a SID. (B, C and D) ZEB-1/δEF1 and ZEB-2/SIP1 bind to activated R-Smads. 293T cells were cotransfected with the indicated expression vectors: 3 µg of either Flag-tagged Smad1, Smad2 or Smad3, 4 µg of either myc-tagged full-length human ZEB-1/δEF1 or ZEB-2/SIP1 and 0.8 µg of the corresponding constitutively active hemagglutinin-tagged constitutively active ALKs ALK6 (Q203D) for Smad1 (B) and ALK5 (T204D) for Smad2 and Smad3 (C and D). After 48 h, cells lysates were immunoprecipitated (IP) for Flag-Smads and the co-immunoprecipitated ZEB-1/δEF1 and ZEB-2/SIP1 was detected by western blotting (WB) with 9E10 myc antibody. The input represents 15% of the lysate.
Fig. 1. (A) Schematic representation of the ZEB family (ZEB-1/δEF1 and ZEB-2/SIP1) of two-handed zinc finger factors. The central region (CR) in between both zinc finger clusters act as a repressor domain in part through the recruitment of the CtBP corepressor through a CID. 3′ of the N-terminal zinc fingers there is a region that acts as a SID. (B, C and D) ZEB-1/δEF1 and ZEB-2/SIP1 bind to activated R-Smads. 293T cells were cotransfected with the indicated expression vectors: 3 µg of either Flag-tagged Smad1, Smad2 or Smad3, 4 µg of either myc-tagged full-length human ZEB-1/δEF1 or ZEB-2/SIP1 and 0.8 µg of the corresponding constitutively active hemagglutinin-tagged constitutively active ALKs ALK6 (Q203D) for Smad1 (B) and ALK5 (T204D) for Smad2 and Smad3 (C and D). After 48 h, cells lysates were immunoprecipitated (IP) for Flag-Smads and the co-immunoprecipitated ZEB-1/δEF1 and ZEB-2/SIP1 was detected by western blotting (WB) with 9E10 myc antibody. The input represents 15% of the lysate.
Fig. 1. (A) Schematic representation of the ZEB family (ZEB-1/δEF1 and ZEB-2/SIP1) of two-handed zinc finger factors. The central region (CR) in between both zinc finger clusters act as a repressor domain in part through the recruitment of the CtBP corepressor through a CID. 3′ of the N-terminal zinc fingers there is a region that acts as a SID. (B, C and D) ZEB-1/δEF1 and ZEB-2/SIP1 bind to activated R-Smads. 293T cells were cotransfected with the indicated expression vectors: 3 µg of either Flag-tagged Smad1, Smad2 or Smad3, 4 µg of either myc-tagged full-length human ZEB-1/δEF1 or ZEB-2/SIP1 and 0.8 µg of the corresponding constitutively active hemagglutinin-tagged constitutively active ALKs ALK6 (Q203D) for Smad1 (B) and ALK5 (T204D) for Smad2 and Smad3 (C and D). After 48 h, cells lysates were immunoprecipitated (IP) for Flag-Smads and the co-immunoprecipitated ZEB-1/δEF1 and ZEB-2/SIP1 was detected by western blotting (WB) with 9E10 myc antibody. The input represents 15% of the lysate.
Fig. 1. (A) Schematic representation of the ZEB family (ZEB-1/δEF1 and ZEB-2/SIP1) of two-handed zinc finger factors. The central region (CR) in between both zinc finger clusters act as a repressor domain in part through the recruitment of the CtBP corepressor through a CID. 3′ of the N-terminal zinc fingers there is a region that acts as a SID. (B, C and D) ZEB-1/δEF1 and ZEB-2/SIP1 bind to activated R-Smads. 293T cells were cotransfected with the indicated expression vectors: 3 µg of either Flag-tagged Smad1, Smad2 or Smad3, 4 µg of either myc-tagged full-length human ZEB-1/δEF1 or ZEB-2/SIP1 and 0.8 µg of the corresponding constitutively active hemagglutinin-tagged constitutively active ALKs ALK6 (Q203D) for Smad1 (B) and ALK5 (T204D) for Smad2 and Smad3 (C and D). After 48 h, cells lysates were immunoprecipitated (IP) for Flag-Smads and the co-immunoprecipitated ZEB-1/δEF1 and ZEB-2/SIP1 was detected by western blotting (WB) with 9E10 myc antibody. The input represents 15% of the lysate.
Fig. 2. (A–C) Interaction of ZEB proteins with Smad1, Smad2 and Smad3 requires TGFβ signaling. Experiments were performed as in Figure 1B–D, but in the absence of the corresponding constitutively active ALKs ALK5 (T204D) and ALK6 (Q203D). (D) Interaction between endogenous ZEB-1/δEF1 and Smad2 and Smad3. Cell lysates from 293T cells immunoprecipitated with either an antibody against ZEB-1/δEF1 or a control antibody (goat-Ig) in the presence or absence of 20 µg of an expression vector for ALK5 (T204D). Proteins immunoprecipitated by ZEB-1/δEF1 were then western blotted for associated endogenous Smad2 and Smad3. NS indicates a non-specific band. (E) The same region in ZEB-1/δEF1 and ZEB-2/SIP1 (Smad-interacting region, SID) mediates their binding to activated Smad1 and Smad3. The indicated myc-tagged constructs for ZEB-1/δEF1 and ZEB-2/SIP1 (see Materials and methods for further details) were tested for their ability to interact with Smad1 and Smad3 in the presence of either constitutively active ALK6 (Q203D) and ALK5 (T204D), respectively.
Fig. 2. (A–C) Interaction of ZEB proteins with Smad1, Smad2 and Smad3 requires TGFβ signaling. Experiments were performed as in Figure 1B–D, but in the absence of the corresponding constitutively active ALKs ALK5 (T204D) and ALK6 (Q203D). (D) Interaction between endogenous ZEB-1/δEF1 and Smad2 and Smad3. Cell lysates from 293T cells immunoprecipitated with either an antibody against ZEB-1/δEF1 or a control antibody (goat-Ig) in the presence or absence of 20 µg of an expression vector for ALK5 (T204D). Proteins immunoprecipitated by ZEB-1/δEF1 were then western blotted for associated endogenous Smad2 and Smad3. NS indicates a non-specific band. (E) The same region in ZEB-1/δEF1 and ZEB-2/SIP1 (Smad-interacting region, SID) mediates their binding to activated Smad1 and Smad3. The indicated myc-tagged constructs for ZEB-1/δEF1 and ZEB-2/SIP1 (see Materials and methods for further details) were tested for their ability to interact with Smad1 and Smad3 in the presence of either constitutively active ALK6 (Q203D) and ALK5 (T204D), respectively.
Fig. 2. (A–C) Interaction of ZEB proteins with Smad1, Smad2 and Smad3 requires TGFβ signaling. Experiments were performed as in Figure 1B–D, but in the absence of the corresponding constitutively active ALKs ALK5 (T204D) and ALK6 (Q203D). (D) Interaction between endogenous ZEB-1/δEF1 and Smad2 and Smad3. Cell lysates from 293T cells immunoprecipitated with either an antibody against ZEB-1/δEF1 or a control antibody (goat-Ig) in the presence or absence of 20 µg of an expression vector for ALK5 (T204D). Proteins immunoprecipitated by ZEB-1/δEF1 were then western blotted for associated endogenous Smad2 and Smad3. NS indicates a non-specific band. (E) The same region in ZEB-1/δEF1 and ZEB-2/SIP1 (Smad-interacting region, SID) mediates their binding to activated Smad1 and Smad3. The indicated myc-tagged constructs for ZEB-1/δEF1 and ZEB-2/SIP1 (see Materials and methods for further details) were tested for their ability to interact with Smad1 and Smad3 in the presence of either constitutively active ALK6 (Q203D) and ALK5 (T204D), respectively.
Fig. 2. (A–C) Interaction of ZEB proteins with Smad1, Smad2 and Smad3 requires TGFβ signaling. Experiments were performed as in Figure 1B–D, but in the absence of the corresponding constitutively active ALKs ALK5 (T204D) and ALK6 (Q203D). (D) Interaction between endogenous ZEB-1/δEF1 and Smad2 and Smad3. Cell lysates from 293T cells immunoprecipitated with either an antibody against ZEB-1/δEF1 or a control antibody (goat-Ig) in the presence or absence of 20 µg of an expression vector for ALK5 (T204D). Proteins immunoprecipitated by ZEB-1/δEF1 were then western blotted for associated endogenous Smad2 and Smad3. NS indicates a non-specific band. (E) The same region in ZEB-1/δEF1 and ZEB-2/SIP1 (Smad-interacting region, SID) mediates their binding to activated Smad1 and Smad3. The indicated myc-tagged constructs for ZEB-1/δEF1 and ZEB-2/SIP1 (see Materials and methods for further details) were tested for their ability to interact with Smad1 and Smad3 in the presence of either constitutively active ALK6 (Q203D) and ALK5 (T204D), respectively.
Fig. 2. (A–C) Interaction of ZEB proteins with Smad1, Smad2 and Smad3 requires TGFβ signaling. Experiments were performed as in Figure 1B–D, but in the absence of the corresponding constitutively active ALKs ALK5 (T204D) and ALK6 (Q203D). (D) Interaction between endogenous ZEB-1/δEF1 and Smad2 and Smad3. Cell lysates from 293T cells immunoprecipitated with either an antibody against ZEB-1/δEF1 or a control antibody (goat-Ig) in the presence or absence of 20 µg of an expression vector for ALK5 (T204D). Proteins immunoprecipitated by ZEB-1/δEF1 were then western blotted for associated endogenous Smad2 and Smad3. NS indicates a non-specific band. (E) The same region in ZEB-1/δEF1 and ZEB-2/SIP1 (Smad-interacting region, SID) mediates their binding to activated Smad1 and Smad3. The indicated myc-tagged constructs for ZEB-1/δEF1 and ZEB-2/SIP1 (see Materials and methods for further details) were tested for their ability to interact with Smad1 and Smad3 in the presence of either constitutively active ALK6 (Q203D) and ALK5 (T204D), respectively.
Fig. 3. ZEB-1/δEF1 synergizes with TGFβ/BMP in transcriptional activation, whereas ZEB-2/SIP1 represses. (A and B) Mv1Lu or HaCAT cells (similar results were obtained in both cell types) were cotransfected with 0.3 µg of a firefly luciferase reporter containing the indicated promoter along with either 0.48 µg of the empty vector (CS2MT, not shown) or 0.7 µg of either ZEB-1/δEF1 or ZEB-2/SIP1 expression vectors and stimulated with TGFβ1 (25 pM for Mv1Lu or 100 pM for HaCAT). (C) C2C12 cells were transfected as in (A) and (B) with a firefly luciferase reporter containing the BMP-2-responsive Xvent2 promoter. Where indicated, cells were treated with 100 ng/ml BMP-2. (D) Regulation of TGFβ-mediated transcription by ZEB proteins is dependent on the transcriptional activity of R-Smads and requires the SID. Mv1Lu or HaCAT cells were transfected with 0.3 µg of 3TPluc and either 0.48 µg of vector (CS2MT, not shown), 0.6 µg of ZEB-1 (ΔSID) or ZEB-2 (ΔSID) (lacking the SID) or 0.7 µg of full-length ZEB-1 or ZEB-2. Where indicated, an expression vector for Smad3-dominant negative (Smad3-DN, D470E) was cotransfected. (E and F) Regulation of TGFβ-mediated transcription by ZEB proteins requires the presence of Smad4. Smad4 (+/+) HCT 116 cells (E) and Smad 4 (–/–) HCT 116/518 cells (F) were cotransfected as in (A) with 3TP-luc, either ZEB-1 or ZEB-2 and ALK5 (T204D) to activate the TGFβ signaling pathway.
Fig. 3. ZEB-1/δEF1 synergizes with TGFβ/BMP in transcriptional activation, whereas ZEB-2/SIP1 represses. (A and B) Mv1Lu or HaCAT cells (similar results were obtained in both cell types) were cotransfected with 0.3 µg of a firefly luciferase reporter containing the indicated promoter along with either 0.48 µg of the empty vector (CS2MT, not shown) or 0.7 µg of either ZEB-1/δEF1 or ZEB-2/SIP1 expression vectors and stimulated with TGFβ1 (25 pM for Mv1Lu or 100 pM for HaCAT). (C) C2C12 cells were transfected as in (A) and (B) with a firefly luciferase reporter containing the BMP-2-responsive Xvent2 promoter. Where indicated, cells were treated with 100 ng/ml BMP-2. (D) Regulation of TGFβ-mediated transcription by ZEB proteins is dependent on the transcriptional activity of R-Smads and requires the SID. Mv1Lu or HaCAT cells were transfected with 0.3 µg of 3TPluc and either 0.48 µg of vector (CS2MT, not shown), 0.6 µg of ZEB-1 (ΔSID) or ZEB-2 (ΔSID) (lacking the SID) or 0.7 µg of full-length ZEB-1 or ZEB-2. Where indicated, an expression vector for Smad3-dominant negative (Smad3-DN, D470E) was cotransfected. (E and F) Regulation of TGFβ-mediated transcription by ZEB proteins requires the presence of Smad4. Smad4 (+/+) HCT 116 cells (E) and Smad 4 (–/–) HCT 116/518 cells (F) were cotransfected as in (A) with 3TP-luc, either ZEB-1 or ZEB-2 and ALK5 (T204D) to activate the TGFβ signaling pathway.
Fig. 3. ZEB-1/δEF1 synergizes with TGFβ/BMP in transcriptional activation, whereas ZEB-2/SIP1 represses. (A and B) Mv1Lu or HaCAT cells (similar results were obtained in both cell types) were cotransfected with 0.3 µg of a firefly luciferase reporter containing the indicated promoter along with either 0.48 µg of the empty vector (CS2MT, not shown) or 0.7 µg of either ZEB-1/δEF1 or ZEB-2/SIP1 expression vectors and stimulated with TGFβ1 (25 pM for Mv1Lu or 100 pM for HaCAT). (C) C2C12 cells were transfected as in (A) and (B) with a firefly luciferase reporter containing the BMP-2-responsive Xvent2 promoter. Where indicated, cells were treated with 100 ng/ml BMP-2. (D) Regulation of TGFβ-mediated transcription by ZEB proteins is dependent on the transcriptional activity of R-Smads and requires the SID. Mv1Lu or HaCAT cells were transfected with 0.3 µg of 3TPluc and either 0.48 µg of vector (CS2MT, not shown), 0.6 µg of ZEB-1 (ΔSID) or ZEB-2 (ΔSID) (lacking the SID) or 0.7 µg of full-length ZEB-1 or ZEB-2. Where indicated, an expression vector for Smad3-dominant negative (Smad3-DN, D470E) was cotransfected. (E and F) Regulation of TGFβ-mediated transcription by ZEB proteins requires the presence of Smad4. Smad4 (+/+) HCT 116 cells (E) and Smad 4 (–/–) HCT 116/518 cells (F) were cotransfected as in (A) with 3TP-luc, either ZEB-1 or ZEB-2 and ALK5 (T204D) to activate the TGFβ signaling pathway.
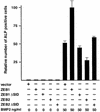
Fig. 4. ZEB-1/δEF1 augments BMP-2-dependent osteogenic differentiation of mesenchymal cells. C2C12 were cotransfected with 8 µg of either vector alone (CS2MT) or expression vectors for ZEB-1, ZEB-2 or their ΔSID deletional mutants (ZEB-1 ΔSID and ZEB2 ΔSID) and treated with the indicated amount of BMP-2. After 4 days, cells were assessed for ALP activity as described in Materials and methods.
Fig. 5. ZEB-1/δEF1 synergizes with TGFβ to mediate G1 cell cycle arrest. (A) Mv1Lu cells were transfected with 20 µg of the empty vector or ZEB-1/δEF1 along with 1 µg of an expression vector for EGFP (as a marker for sorting transfected cells) and, where indicated, treated with either 18 pM (suboptimal dose) or 75 pM (maximum growth arrest dose) TGFβ1. The cell cycle profile of EGFP-positive cells was analyzed by FACS analysis. (B) As in (A), but cells were preincubated for 48 h with 75 ng/ml nocodazole to induce G2/M arrest (see Materials and methods and Zhang et al., 2000). (C) Mv1Lu cells were transfected with 20 µg of either empty vector, full-length ZEB-1/δEF1 or a mutant of ZEB-1/δEF1 with a deletion of the SID (ZEB-1/δEF1 ΔSID) and EGFP in the presence of nocodazole as in (B). The percentage of cells in G1 phase was determined by FACS and their value compared in the absence or presence of TGFβ1 (both suboptimal and maximum effect doses).
Fig. 5. ZEB-1/δEF1 synergizes with TGFβ to mediate G1 cell cycle arrest. (A) Mv1Lu cells were transfected with 20 µg of the empty vector or ZEB-1/δEF1 along with 1 µg of an expression vector for EGFP (as a marker for sorting transfected cells) and, where indicated, treated with either 18 pM (suboptimal dose) or 75 pM (maximum growth arrest dose) TGFβ1. The cell cycle profile of EGFP-positive cells was analyzed by FACS analysis. (B) As in (A), but cells were preincubated for 48 h with 75 ng/ml nocodazole to induce G2/M arrest (see Materials and methods and Zhang et al., 2000). (C) Mv1Lu cells were transfected with 20 µg of either empty vector, full-length ZEB-1/δEF1 or a mutant of ZEB-1/δEF1 with a deletion of the SID (ZEB-1/δEF1 ΔSID) and EGFP in the presence of nocodazole as in (B). The percentage of cells in G1 phase was determined by FACS and their value compared in the absence or presence of TGFβ1 (both suboptimal and maximum effect doses).
Fig. 5. ZEB-1/δEF1 synergizes with TGFβ to mediate G1 cell cycle arrest. (A) Mv1Lu cells were transfected with 20 µg of the empty vector or ZEB-1/δEF1 along with 1 µg of an expression vector for EGFP (as a marker for sorting transfected cells) and, where indicated, treated with either 18 pM (suboptimal dose) or 75 pM (maximum growth arrest dose) TGFβ1. The cell cycle profile of EGFP-positive cells was analyzed by FACS analysis. (B) As in (A), but cells were preincubated for 48 h with 75 ng/ml nocodazole to induce G2/M arrest (see Materials and methods and Zhang et al., 2000). (C) Mv1Lu cells were transfected with 20 µg of either empty vector, full-length ZEB-1/δEF1 or a mutant of ZEB-1/δEF1 with a deletion of the SID (ZEB-1/δEF1 ΔSID) and EGFP in the presence of nocodazole as in (B). The percentage of cells in G1 phase was determined by FACS and their value compared in the absence or presence of TGFβ1 (both suboptimal and maximum effect doses).
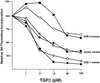
Fig. 6. ZEB proteins regulate TGFβ-dependent growth suppression. Mv1Lu cells were stably transfected with either the empty vector, ZEB-1/δEF1 or ZEB-2/SIP1. Stable clones were treated with different doses of TGFβ1 and assessed for cell proliferation by measuring their [3H]thymidine uptake. Two representative clones for each construct are shown.
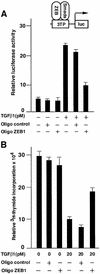
Fig. 7. Endogenous ZEB/δEF1 mediates TGFβ functions. (A) Endogenous ZEB/δEF1 is important for TGFβ-dependent transcriptional activation. Mv1Lu cells were transfected with 0.3 µg of the TGFβ-responsive 3TP promoter and transcriptional activity assessed as in Figure 3. Where indicated, either antisense oligos to ZEB-1/δEF1 (Oligo ZEB1) or a scrambled control oligo (Oligo control) were added (see Materials and methods). One-tenth of a microgram of SV40βgal was cotransfected to control for transfection efficiency. (B) Endogenous ZEB/δEF1 is important for TGFβ-mediated growth suppression. Mv1Lu cells were transfected with antisense oligos and 0.2 µg of puroBabe, and treated with 1 mg/ml puromycin and 20 pM TGFβ1 for 36 h before the proliferation was assessed by the incorporation of [3H]thymidine.
Similar articles
- Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins.
Postigo AA, Depp JL, Taylor JJ, Kroll KL. Postigo AA, et al. EMBO J. 2003 May 15;22(10):2453-62. doi: 10.1093/emboj/cdg226. EMBO J. 2003. PMID: 12743039 Free PMC article. - SIP1 (Smad interacting protein 1) and deltaEF1 (delta-crystallin enhancer binding factor) are structurally similar transcriptional repressors.
van Grunsven LA, Schellens A, Huylebroeck D, Verschueren K. van Grunsven LA, et al. J Bone Joint Surg Am. 2001;83-A Suppl 1(Pt 1):S40-7. J Bone Joint Surg Am. 2001. PMID: 11263664 Review. - deltaEF1 represses BMP-2-induced differentiation of C2C12 myoblasts into the osteoblast lineage.
Yang S, Zhao L, Yang J, Chai D, Zhang M, Zhang J, Ji X, Zhu T. Yang S, et al. J Biomed Sci. 2007 Sep;14(5):663-79. doi: 10.1007/s11373-007-9155-5. Epub 2007 May 4. J Biomed Sci. 2007. PMID: 17479358 - OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways.
Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massagué J. Hata A, et al. Cell. 2000 Jan 21;100(2):229-40. doi: 10.1016/s0092-8674(00)81561-5. Cell. 2000. PMID: 10660046 - Transforming growth factor beta signalling in vitro and in vivo: activin ligand-receptor interaction, Smad5 in vasculogenesis, and repression of target genes by the deltaEF1/ZEB-related SIP1 in the vertebrate embryo.
Zwijsen A, van Grunsven LA, Bosman EA, Collart C, Nelles L, Umans L, Van de Putte T, Wuytens G, Huylebroeck D, Verschueren K. Zwijsen A, et al. Mol Cell Endocrinol. 2001 Jun 30;180(1-2):13-24. doi: 10.1016/s0303-7207(01)00505-6. Mol Cell Endocrinol. 2001. PMID: 11451567 Review.
Cited by
- Selective activation of p120ctn-Kaiso signaling to unlock contact inhibition of ARPE-19 cells without epithelial-mesenchymal transition.
Chen HC, Zhu YT, Chen SY, Tseng SC. Chen HC, et al. PLoS One. 2012;7(5):e36864. doi: 10.1371/journal.pone.0036864. Epub 2012 May 9. PLoS One. 2012. PMID: 22590627 Free PMC article. - Intrinsic Balance between ZEB Family Members Is Important for Melanocyte Homeostasis and Melanoma Progression.
Bruneel K, Verstappe J, Vandamme N, Berx G. Bruneel K, et al. Cancers (Basel). 2020 Aug 11;12(8):2248. doi: 10.3390/cancers12082248. Cancers (Basel). 2020. PMID: 32796736 Free PMC article. Review. - Zinc finger proteins in cancer progression.
Jen J, Wang YC. Jen J, et al. J Biomed Sci. 2016 Jul 13;23(1):53. doi: 10.1186/s12929-016-0269-9. J Biomed Sci. 2016. PMID: 27411336 Free PMC article. Review. - Epigenetic Regulation of Inflammatory Cytokine-Induced Epithelial-To-Mesenchymal Cell Transition and Cancer Stem Cell Generation.
Markopoulos GS, Roupakia E, Marcu KB, Kolettas E. Markopoulos GS, et al. Cells. 2019 Sep 25;8(10):1143. doi: 10.3390/cells8101143. Cells. 2019. PMID: 31557902 Free PMC article. Review. - Emerging mechanisms progress of colorectal cancer liver metastasis.
Zhao W, Dai S, Yue L, Xu F, Gu J, Dai X, Qian X. Zhao W, et al. Front Endocrinol (Lausanne). 2022 Dec 8;13:1081585. doi: 10.3389/fendo.2022.1081585. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36568117 Free PMC article. Review.
References
- Attisano L. and Wrana,J.L. (2000) Smads as transcriptional modulators. Curr. Opin. Cell Biol., 12, 235–243. - PubMed
- Berry F.B., Miura,Y. Mihara,K., Kaspar,P., Sakata,N., Hashimoto-Tamaoki,T. and Tamaoki,T. (2001) Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J. Biol. Chem., 276, 25057–25065. - PubMed
- Brabletz T., Jung,A., Hlubek,F., Lohberg,C., Meiler,J., Suchy,U. and Kirchner,T. (1999) Negative regulation of CD4 expression in T cells by the transcriptional repressor ZEB. Int. Immunol., 11, 1701–1708. - PubMed
- Broihier H.T, Moore,L.A., Van Doren,M., Newman,S. and Lehmann,R. (1998) zfh-1 is required for germ cell migration and gonadal mesoderm development in Drosophila. Development, 125, 655–666. - PubMed
- Cacheux V., Dastot-Le Moal,F., Kaariainen,H., Bondurand,N., Rintala,R., Boissier,B., Wilson,M., Mowat,D. and Goossens,M. (2001) Loss-of-function mutations in SIP1 (Smad interacting protein 1) result in a syndromic Hirschsprung disease. Hum. Mol. Genet., 10, 1503–1510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials