A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity - PubMed (original) (raw)
A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity
Georg Lipps et al. EMBO J. 2003.
Abstract
Although DNA replication is a process common in all domains of life, primase and replicative DNA polymerase appear to have evolved independently in the bacterial domain versus the archaeal/eukaryal branch of life. Here, we report on a new type of replication protein that constitutes the first member of the DNA polymerase family E. The protein ORF904, encoded by the plasmid pRN1 from the thermoacidophile archaeon Sulfolobus islandicus, is a highly compact multifunctional enzyme with ATPase, primase and DNA polymerase activity. Recombinant purified ORF904 hydrolyses ATP in a DNA-dependent manner. Deoxynucleotides are preferentially used for the synthesis of primers approximately 8 nucleotides long. The DNA polymerase activity of ORF904 synthesizes replication products of up to several thousand nucleotides in length. The primase and DNA polymerase activity are located in the N-terminal half of the protein, which does not show homology to any known DNA polymerase or primase. ORF904 constitutes a new type of replication enzyme, which could have evolved independently from the eubacterial and archaeal/eukaryal proteins of DNA replication.
Figures
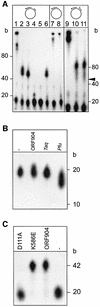
Fig. 1. Hypothetical domain organization and purification of ORF904. (A) Two domains can be identified by sequence comparison: an N-terminal domain, presumably carrying the primase and DNA polymerase activity; and a C-terminal helicase domain. The N-terminal domain was tentatively named prim/pol domain. The middle part of ORF904 has no sequence similarity to known proteins. ORF904 is, however, conserved within the pRN plasmid family, and ORF904 homologues are also found integrated into the genomes of S.tokodaii and S.solfataricus. Small vertical bars indicate the position of the two point mutations D111A and K586E. The bacteriophage P4 α protein has a Toprim domain (Aravind et al., 1998) and a super family III helicase domain (Gorbalenya et al., 1990). An uncharacterized S.coelicolor protein is the only protein that also has the prim/pol domain and the helicase domain in one polypeptide chain. These two domains are encoded by two adjacent genes in the case of bacteriophages Sfi21 and Phi31. The expect values of BLASTP (Altschul et al., 1990) indicate the sequence conservation of the respective domain towards ORF904. Proteins and domains are drawn to scale with the length of the proteins indicated by the number of amino acids. (B) Alignment of the putative prim/pol domains. Included into the alignment by Multalign (Corpet, 1988) are three bacterial sequences only distantly related to the archaeal plasmidal proteins encoded by pRN1 and pRN2. Conserved amino acids are shown in red letters. Conserved acid residues, which are candidates for active site residues, are boxed. Numbers indicate the length of weakly conserved amino acid stretches, which have been omitted for clarity. The consensus symbols are ! (IV), $ (LM), % (FY) and # (NDQE). (C) Purification of ORF904. Coomassie Blue stain of a protein gel: extracts from uninduced and induced E.coli BL21 codon plus cells are shown in lanes 1 and 2. Recombinant His-tagged ORF904 was purified by cobalt chelate chromatography (flow-through, lane 3; pool, lane 4) and sulphate cation-exchange chromatography (flow-through, lane 5; pool, lane 6).
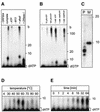
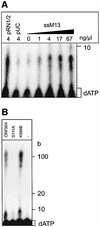
Fig. 2. ATPase activity of ORF904. (A) ORF904 from pRN1 and the homologous proteins from the other pRN plasmids contain a complete Walker A motif. Residues corresponding to the Walker A consensus motif are in bold. K586E denotes the mutated Walker A motif. (B) ORF904 (0.6 µM) was incubated with [γ-32P]ATP and 4 ng/µl nucleic acids for 10 min at 80°C. The basal ATPase activity of ORF904 is very low (lane control) but there is significant stimulation by the double-stranded DNA pUC19 and pRN1/2. Single-stranded DNA is a weaker activator of the ATPase activity (lane ssM13), and bulk tRNA from yeast (lane tRNA) does not stimulate ORF904. No ATPase activity is seen without ORF904 (lane -ORF904). (C) Kinetics of ATP hydrolysis. The reactions were carried out in the presence of 3 mM ATP and 0.6 µM ORF904 in a volume of 30 µl. ORF904 was tested in the presence of double-stranded DNA (filled circles) and absence of DNA (open circles). The mutant protein K586E (triangle) was also assayed in the presence of DNA. (D) Concentration dependence of ATP hydrolysis measured at saturating DNA concentrations (40 ng/µl). The data were fitted according to Michaelis–Menten kinetics. Error bars indicate standard deviations from several independent determinations. (E) ATPase activity of the mutant proteins. ATP hydrolysis by 0.4 µM protein was assayed in the presence and absence of 40 ng/µl double-stranded DNA. No ATPase activity is seen for the Walker A mutant K586E. The DNA-dependent ATPase activity of the mutant D111A is unaffected.
Fig. 3. ORF904 is a DNA polymerase. (A) ORF904 (0.4 µM) was incubated with a short primer–template substrate (CGAACCCGTT CTCGGAGCAC hybridized to TTCTGCACAAAGCGGTTCTGCAG TGCTCCGAGAACGGGTTCG). Primers are extended in the presence of 0.2 mM dNTPs (lane dNTP) during 10 min incubation at 50°C. In control reactions with 0.2 mM rNTPs (lane rNTPs) or without template (data not shown), no primer elongation is observable. (B) The primer extension activity of ORF904 was assayed between 50 and 90°C. A 5′-32P-labelled 30 nt primer hybridized onto M13 DNA was incubated with 0.4 µM ORF904, 0.2 mM dNTPs, 0.2 mM ATP for 15 min at the given temperatures. Extension products were analysed on a 5% denaturing polyacrylamide gel. (C) The kinetics of primer extension was followed at 65°C. ORF904 (0.4 µM) was incubated with single-primed M13 in the presence of 0.2 mM dNTPs and 0.2 mM ATP.
Fig. 4. ORF904 has no exonuclease activity and D111A has no DNA polymerase activity. (A) ORF904 (0.4 µM) was incubated with a double (lanes 1–6 and 9–11) and single (lanes 7 and 8) primed M13 DNA. The upstream oligodeoxynucleotide of the double-primed substrates was either blunt ended (lanes 1–6) or had a 15 nuncleotide 5′-tail (lanes 9–11). Taq polymerase (0.01 U/µl) was used for primer extension in lanes 1, 7 and 9; in all other lanes, 0.4 µM ORF904 was included. The following nucleotides were added: 0.2 mM dNTP (lanes 1, 2, 7, 8 and 10), 0.2 mM rNTPs and 0.2 mM dNTPs (lane 3), 0.2 mM rNTPs (lane 4), no nucleotides (lane 5), 0.2 mM dNTPs and 1 mM ATP (lanes 6 and 11 ). In contrast to Taq polymerase, ORF904 is not able to extend the polymerization past the beginning of the second more upstream primer. Some albeit weak polymerization past the beginning of the upstream oligodeoxynucleotide (marked with an arrow) is seen when the upstream oligodeoxynucleotide is 5′-tailed (lanes 10 and 11). (B) A 5′-end-labelled primer–template substrate (see Figure 3A) with a mismatch at the 3′-end of the primer was incubated in the absence of dNTPs for 30 min at 65°C. Lane –, no enzyme added; lane ORF904, 0.4 µM ORF904; lane Taq, 0.02 U/µl Taq polymerase; lane Pfu, 0.02 U/µl Pfu polymerase. The reaction products were analysed by denaturing PAGE. ORF904 does not degrade the primer whereas Pfu polymerase does. (C) DNA polymerase activity of the mutants. Wild-type ORF904 and the mutant K586E are able to extend the primer of the short primer–template substrate. D111A has no detectable DNA polymerase activity.
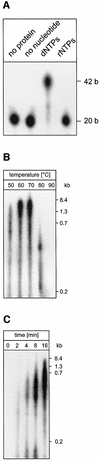
Fig. 5. ORF904 synthesizes and elongates a primer. (A) M13 single-stranded DNA as template: 0.2 µM ORF904 was incubated for 30 min at 50°C with 10 µM rNTPs in the presence of [α-32P]ATP. A primer is only observed when 0.6 g/l single-stranded M13 is present and alkali treatment is omitted. (B) pRN1/2 as template: 0.04 g/l of pRN1/2 was assayed as in (A). The primer (lane 1) can be extended to longer products with a further 30 min incubation in the presence of 0.2 mM dNTPs (lane 2) or of 0.2 mM dNTPs and 0.5 U Taq polymerase (lane 3). Neither primer nor extension products are seen when ORF904 is omitted from the reaction with Taq polymerase (lane 4).
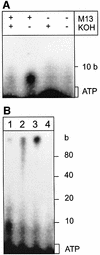
Fig. 6. ORF904 preferentially incorporates dNTPs into primers. (A) The complete reaction mix contained 0.2 µM ORF904, 10 µM dNTPs, [α-32P]dATP, 0.03 g/l single-stranded M13 DNA and 1 mM ATP, and was incubated for 30 min at 50°C. The last two reactions were treated with 0.3 M KOH and 2 U of DNAse I, respectively. As DNase I digests DNA to oligodeoxynucleotides, a smear of short products is visible in lane DNase I. (B) As in (A), but with [α-32P]dGTP as label. Varying amounts of ATP and non-hydrolysable analogues of ATP were used. (C) Determination of primer length. Lane P, products of primase reaction; lane M, 10 base ladder. (D) The primase activity of ORF904 was assayed between 4 and 90°C for 30 min (reaction conditions as in A). (E) The time course of a primase reaction at 50°C is shown (reaction conditions as in A).
Fig. 7. The native double-stranded plasmids can be primed by ORF904. Primase activity of the mutants D111A and K586E. (A) Different substrates were used in priming reactions in the presence of 10 µM rNTPs and [α-32P]dATP. Whereas double-stranded DNA pUC19 DNA was not primed, the positively supercoiled mixture of plasmids pRN1 and pRN2 isolated from S.islandicus was more efficiently primed than the same amount of single-stranded M13 DNA. A pUC derivate containing the complete sequence of pRN1 is not primed (data not shown). (B) Primase activity of wild-type ORF904 and the mutants K586E and D111A (reaction conditions as in Figure 6A). Mutant D111A is deficient in primase activity.
Similar articles
- Structure of a bifunctional DNA primase-polymerase.
Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Lipps G, et al. Nat Struct Mol Biol. 2004 Feb;11(2):157-62. doi: 10.1038/nsmb723. Epub 2004 Jan 18. Nat Struct Mol Biol. 2004. PMID: 14730355 - Molecular modeling and functional characterization of the monomeric primase-polymerase domain from the Sulfolobus solfataricus plasmid pIT3.
Prato S, Vitale RM, Contursi P, Lipps G, Saviano M, Rossi M, Bartolucci S. Prato S, et al. FEBS J. 2008 Sep;275(17):4389-402. doi: 10.1111/j.1742-4658.2008.06585.x. Epub 2008 Jul 30. FEBS J. 2008. PMID: 18671730 - Properties of an unusual DNA primase from an archaeal plasmid.
Beck K, Lipps G. Beck K, et al. Nucleic Acids Res. 2007;35(17):5635-45. doi: 10.1093/nar/gkm625. Epub 2007 Aug 20. Nucleic Acids Res. 2007. PMID: 17709343 Free PMC article. - The replication protein of the Sulfolobus islandicus plasmid pRN1.
Lipps G. Lipps G. Biochem Soc Trans. 2004 Apr;32(Pt 2):240-4. doi: 10.1042/bst0320240. Biochem Soc Trans. 2004. PMID: 15046580 Review. - The Pol α-primase complex.
Pellegrini L. Pellegrini L. Subcell Biochem. 2012;62:157-69. doi: 10.1007/978-94-007-4572-8_9. Subcell Biochem. 2012. PMID: 22918585 Review.
Cited by
- Novel RepA-MCM proteins encoded in plasmids pTAU4, pORA1 and pTIK4 from Sulfolobus neozealandicus.
Greve B, Jensen S, Phan H, Brügger K, Zillig W, She Q, Garrett RA. Greve B, et al. Archaea. 2005 May;1(5):319-25. doi: 10.1155/2005/159218. Archaea. 2005. PMID: 15876565 Free PMC article. - Human Primpol1: a novel guardian of stalled replication forks.
Im JS, Lee KY, Dillon LW, Dutta A. Im JS, et al. EMBO Rep. 2013 Dec;14(12):1032-3. doi: 10.1038/embor.2013.171. Epub 2013 Nov 5. EMBO Rep. 2013. PMID: 24189099 Free PMC article. No abstract available. - A four-in-one replicase integrating key enzymatic activities for DNA replication.
Zhang Y, Lu X, Zhu B, Huang F. Zhang Y, et al. Nucleic Acids Res. 2025 Jun 20;53(12):gkaf542. doi: 10.1093/nar/gkaf542. Nucleic Acids Res. 2025. PMID: 40548937 Free PMC article. - Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea.
Berkner S, Grogan D, Albers SV, Lipps G. Berkner S, et al. Nucleic Acids Res. 2007;35(12):e88. doi: 10.1093/nar/gkm449. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576673 Free PMC article. - Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus.
Lipps G. Lipps G. Extremophiles. 2006 Feb;10(1):17-28. doi: 10.1007/s00792-005-0492-x. Epub 2006 Jan 6. Extremophiles. 2006. PMID: 16397749 Review.
References
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
- Augustin M.A., Huber,R. and Kaiser,J.T. (2001) Crystal structure of a DNA-dependent RNA polymerase (DNA primase). Nat. Struct. Biol., 8, 57–61. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources