Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein - PubMed (original) (raw)
Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein
Gary K Geiss et al. J Virol. 2003 Jun.
Abstract
Alpha/beta interferons (IFN-alpha/beta) induce potent antiviral and antiproliferative responses and are used to treat a wide range of human diseases, including chronic hepatitis C virus (HCV) infection. However, for reasons that remain poorly understood, many HCV isolates are resistant to IFN therapy. To better understand the nature of the cellular IFN response, we examined the effects of IFN treatment on global gene expression by using several types of human cells, including HeLa cells, liver cell lines, and primary fetal hepatocytes. In response to IFN, 50 of the approximately 4,600 genes examined were consistently induced in each of these cell types and another 60 were induced in a cell type-specific manner. A search for IFN-stimulated response elements (ISREs) in genomic DNA located upstream of IFN-stimulated genes revealed both previously identified and novel putative ISREs. To determine whether HCV can alter IFN-regulated gene expression, we performed microarray analyses on IFN-treated HeLa cells expressing the HCV nonstructural 5A (NS5A) protein and on IFN-treated Huh7 cells containing an HCV subgenomic replicon. NS5A partially blocked the IFN-mediated induction of 14 IFN-stimulated genes, an effect that may play a role in HCV resistance to IFN. This block may occur through repression of ISRE-mediated transcription, since NS5A also inhibited the IFN-mediated induction of a reporter gene driven from an ISRE-containing promoter. In contrast, the HCV replicon had very little effect on IFN-regulated gene expression. These differences highlight the importance of comparing results from multiple model systems when investigating complex phenomena such as the cellular response to IFN and viral mechanisms of IFN resistance.
Figures
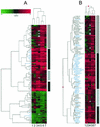
FIG. 1.
IFN induces conserved and cell type-specific changes in gene expression. Lanes 1 and 2, HeLa cells; lanes 3 and 4; Huh7 cells; lanes 5 and 6, primary fetal hepatocytes; lane 7, HH2 cells. (A) Hierarchical clustering of the union set of genes that were differentially regulated in at least one cell type (P ≤ 0.005 for each pair of duplicate experiments). The scale at top indicates the magnitude of induction or repression (log10). Red bars represent genes that were induced by IFN, green bars represent genes that were repressed by IFN, black bars represent genes that were not differentially expressed, and gray bars represent genes that were below the intensity threshold in that experiment. IFN-induced changes in gene expression that were conserved across all cell types are represented by the vertical black bars to the right, primary fetal hepatocyte-specific changes in gene expression are indicated by the vertical hatched bar, and Huh7 cell-specific changes are indicated by the vertical gray bar. A magnified view of genes that were down-regulated by IFN is available at
http://www.expression.washington.edu/public
. (B) Magnified view of ISGs detected by microarray analysis. The symbols for genes that were induced by IFN in all cell types are highlighted in blue.
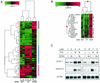
FIG. 2.
NS5A alters IFN-regulated changes in gene expression. (A) Clustering of differentially expressed genes detected in IFN-treated cells in the presence or absence of wild-type or mutant forms of NS5A. Genes induced by IFN and down-regulated by NS5A are indicated by the vertical black bar to the right, genes down-regulated by both forms of NS5A are indicated by the vertical green bar, genes induced by both forms of NS5A are indicated by the vertical red bar, and genes that were regulated differently by NS5A-1B and NS5A-1B5 are indicated by the vertical blue bar. Genes were selected based on a ≥2-fold change and a P value of ≤0.005 in at least two experiments. Lanes 1 and 2, NS5A-1B5 (mutant); lanes 3 to 5, NS5A-1B (wild type); lanes 6 and 7, no NS5A. Each lane represents a unique experiment (and separate RNA isolation) using the cell line indicated. (B) Magnified view of ISGs whose expression was inhibited by NS5A. *, genes with known or putative ISREs (Table 1). Lane designations are the same as described for panel A. (C) Northern blot analysis of STAT1 and ISGF3G (IRF9) mRNAs during IFN treatment of HeLa cells expressing wild-type or mutant NS5A.
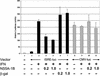
FIG. 3.
NS5A inhibits ISRE-mediated gene transcription. HeLa cells were transfected with plasmid constructs as indicated. Lane 1, pTRE; lanes 2 and 3, pISRE-Luc; lane 4; pISRE-Luc + NS5A (0.2 μg of DNA); lane 5, pISRE-Luc + NS5A (1.0 μg of DNA); lane 6, pISRE-Luc + β-Gal (0.2 μg of DNA); lane 7, pISRE-Luc + β-Gal (1.0 μg of DNA); lanes 8 and 9, pCMV-Luc; lane 10, pCMV-Luc + NS5A (0.2 μg of DNA); lane 11, pCMV-Luc + NS5A (1.0 μg of DNA). Following transfection, cells were mock treated or treated with IFN for 16 h as indicated. Cell lysates were then prepared, and luciferase activity was measured. Luciferase activity is expressed as relative light units (RLU) on the y axis. All experiments were performed in triplicate. Mean values and error bars are shown.
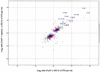
FIG. 4.
IFN-regulated changes in gene expression in Huh7 cells in the presence and absence of an HCV subgenomic replicon are similar. A scatter plot of log2 expression ratios for differentially regulated genes (P ≤ 0.01) in Huh7 cells (x axis) and Huh7 cells harboring the replicon (y axis) in the presence or absence of IFN is shown. Blue crosses represent genes that were differentially expressed in IFN-treated Huh7 cells, magenta crosses represent genes that were differentially expressed in IFN-treated Huh7-replicon cells, and black crosses represent genes that were differentially expressed in both experiments. The ISGs that were down-regulated by NS5A in HeLa cells are highlighted by red circles. (Note that 4 of the 14 ISGs repressed by NS5A in HeLa cells were not induced by IFN in Huh7 cells).
Similar articles
- Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response.
Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR. Polyak SJ, et al. J Virol. 2001 Jul;75(13):6095-106. doi: 10.1128/JVI.75.13.6095-6106.2001. J Virol. 2001. PMID: 11390611 Free PMC article. - Identification of the nonstructural protein 4B of hepatitis C virus as a factor that inhibits the antiviral activity of interferon-alpha.
Xu J, Liu S, Xu Y, Tien P, Gao G. Xu J, et al. Virus Res. 2009 Apr;141(1):55-62. doi: 10.1016/j.virusres.2009.01.001. Epub 2009 Jan 29. Virus Res. 2009. PMID: 19185598 - Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance.
Gale MJ Jr, Korth MJ, Katze MG. Gale MJ Jr, et al. Clin Diagn Virol. 1998 Jul 15;10(2-3):157-62. doi: 10.1016/s0928-0197(98)00034-8. Clin Diagn Virol. 1998. PMID: 9741641 Review. - Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication.
Polyak SJ, Paschal DM, McArdle S, Gale MJ Jr, Moradpour D, Gretch DR. Polyak SJ, et al. Hepatology. 1999 Apr;29(4):1262-71. doi: 10.1002/hep.510290438. Hepatology. 1999. PMID: 10094974
Cited by
- CK2 phosphorylation of CMTR1 promotes RNA cap formation and influenza virus infection.
Lukoszek R, Inesta-Vaquera F, Brett NJM, Liang S, Hepburn LA, Hughes DJ, Pirillo C, Roberts EW, Cowling VH. Lukoszek R, et al. Cell Rep. 2024 Jul 23;43(7):114405. doi: 10.1016/j.celrep.2024.114405. Epub 2024 Jun 25. Cell Rep. 2024. PMID: 38923463 Free PMC article. - CMTR1 is recruited to transcription start sites and promotes ribosomal protein and histone gene expression in embryonic stem cells.
Liang S, Silva JC, Suska O, Lukoszek R, Almohammed R, Cowling VH. Liang S, et al. Nucleic Acids Res. 2022 Mar 21;50(5):2905-2922. doi: 10.1093/nar/gkac122. Nucleic Acids Res. 2022. PMID: 35212377 Free PMC article. - CMTR1-Catalyzed 2'-O-Ribose Methylation Controls Neuronal Development by Regulating Camk2α Expression Independent of RIG-I Signaling.
Lee YL, Kung FC, Lin CH, Huang YS. Lee YL, et al. Cell Rep. 2020 Oct 20;33(3):108269. doi: 10.1016/j.celrep.2020.108269. Cell Rep. 2020. PMID: 33086056 Free PMC article. - Structural analysis, virtual screening and molecular simulation to identify potential inhibitors targeting 2'-O-ribose methyltransferase of SARS-CoV-2 coronavirus.
Jiang Y, Liu L, Manning M, Bonahoom M, Lotvola A, Yang Z, Yang ZQ. Jiang Y, et al. J Biomol Struct Dyn. 2022 Feb;40(3):1331-1346. doi: 10.1080/07391102.2020.1828172. Epub 2020 Oct 4. J Biomol Struct Dyn. 2022. PMID: 33016237 Free PMC article. - Association of the polymorphism of the Toll-like receptor (TLR)-3 and TLR-9 genes with hepatitis C virus-specific cell-mediated immunity outcomes among Egyptian health-care workers.
Abdelwahab SF, Hamdy S, Osman AM, Zakaria ZA, Galal I, Sobhy M, Hashem M, Allam WR, Abdel-Samiee M, Rewisha E, Waked I. Abdelwahab SF, et al. Clin Exp Immunol. 2021 Jan;203(1):3-12. doi: 10.1111/cei.13514. Epub 2020 Oct 2. Clin Exp Immunol. 2021. PMID: 32939755 Free PMC article. Clinical Trial.
References
- Aizaki, H., S. Saito, T. Ogino, N. Miyajima, T. Harada, Y. Matsuura, T. Miyamura, and M. Kohase. 2000. Suppression of interferon-induced antiviral activity in cells expressing hepatitis C virus proteins. J. Interferon Cytokine Res. 20:1111-1120. - PubMed
- Battaglia, A. M., and K. O. Hagmeyer. 2000. Combination therapy with interferon and ribavirin in the treatment of chronic hepatitis C infection. Ann. Pharmacother. 34:487-494. - PubMed
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
- Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. P. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME) toward standards for microarray data. Nat. Genet. 29:365-371. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases