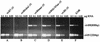Functional subsets of the virB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010 - PubMed (original) (raw)
Functional subsets of the virB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010
Zhenying Liu et al. J Bacteriol. 2003 Jun.
Abstract
The virB-encoded type IV transport complex of Agrobacterium tumefaciens mediates the transfer of DNA and proteins into plant cells, as well as the conjugal transfer of IncQ plasmids, such as RSF1010, between Agrobacterium strains. While several studies have indicated that there are physical interactions among the 11 VirB proteins, the functional significance of the interactions has been difficult to establish since all of the proteins are required for substrate transfer. Our previous studies, however, indicated that although all of the VirB proteins are required for the capacity of a strain to serve as an RSF1010 donor, only a subset of these proteins in the recipient is necessary to increase the conjugal frequency by 3 to 4 logs. The roles of particular groups of VirB proteins in this increased recipient activity were examined in the study reported here. Examination of the expression of subgroups of virB genes revealed that translation of virB6 is necessary for expression of downstream open reading frames. Expression of limited subsets of the VirB proteins in a recipient strain lacking the Ti plasmid revealed that the VirB7 to VirB10 proteins yield a subcomplex that is functional in the recipient assay but that the VirB1 to VirB4 proteins, as a group, dramatically increase this activity in strains expressing VirB7 to VirB10. Finally, the membrane distribution and cross-linking patterns of VirB10, but not of VirB8 or VirB9, in a strain expressing only VirB7 to VirB10 are significantly altered compared to the patterns of the wild type. These characteristics are, however, restored to the wild-type status by coexpression of VirB1 to VirB3. Taken together, these results define subsets of type IV transport complex proteins that are critical in allowing a strain to participate as a recipient in virB-mediated conjugal RSF1010 transfer.
Figures
FIG. 1.
Maps and complementation activities of various virB constructs. Plasmids were constructed as described in Table 1 and the text. The capacity to complement either the PC1006 (virB6 deletion) or PC1008 (virB8 deletion) strain of A. tumefaciens was tested by utilizing the tobacco leaf explant assay system described in Materials and Methods. A representative leaf explant was photographed in each case. The number of tumors per leaf explant (mean ± standard error; n = 14) is indicated below each photograph. In this experiment wild-type strain A348 yielded 11.7 ± 0.8 tumors/leaf explant, and PC1006 and PC1008 yielded no tumors. Wt. wild type; aa, amino acid.
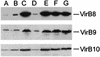
FIG. 2.
Immunoblot analysis of various virB constructs expressed in A136(pAB123). Lane A, pZL36 (virB7 to virB10); lane B, pZL36 and pZL3 (virB7 to virB10, virB1 to virB4); lane C, pZL48 (virB6 to virB10); lane D, pZL48-_Xba_I (virB6 to virB10 with _Xba_I linker insertion in virB6); lane E, pZL48-D2 (virB6 to virB10 with deletion from amino acid 151 to amino acid 237 of VirB6); lane F, pZL48-m12 (virB6 to virB10 with point mutations at amino acids 133 and 134 of VirB6); lane G, A348 (wild-type Ti plasmid).
FIG. 3.
RT-PCR analysis of virB8 transcript levels of Agrobacterium virB6 mutant strains. RT reactions were performed with 0.3, 0.1, and 0.03 μg of RNA isolated from AS-induced cells as described in Materials and Methods. virB1 was used as an internal control during RT-PCR. (A) A136(pAB123, pZL36, pZL3) (virB7 to virB10, virB1 to virB4); (B) A136(pAB123, pZL48-_Xba_I, pZL3) (virB6xbaI-10, virB1 to virB4); (C) A136(pAB123, pZL48-m12, pZL3) (virB6mut-10, virB1 to virB4); (D) A136(pAB123, pZL48-D5, pZL3) (virB6_Δ_185aa-10, virB1 to virB4); (E) A136(pAB123, pZL48, pZL3) (virB6 to virB10, virB1 to virB4); (F) A348.
FIG. 4.
Recipient activities of strain A136(pAB123) carrying plasmids expressing various subsets of the VirB proteins. The adjusted mean numbers of transconjugants per output recipient (from the Tukey-Kramer analysis [see Materials and Methods]) are indicated along with 2 standard errors. An asterisk indicates that a value is significantly different than the value obtained for A136(pAB123) at a 95% confidence level; a number sign indicates that a value is not significantly different from the value obtained for A136 (see Materials and Methods for details concerning statistics).
FIG. 5.
Recipient activities of strain A136(pAB123) containing pZL48 (virB6 to virB10) (A) or pZL48-D2 (virB6_Δ_87aa-10) or pZL48-D5 (virB6_Δ_185aa-10) (B) with or with out other virB genes, as indicated. The mean numbers of transconjugants per output recipient are indicated. The error bars (some of which are not visible due to scale) indicate the standard errors for three separate conjugations performed on the same day. Similar results were observed in two other experiments.
FIG. 6.
Membrane localization of VirB8, VirB9, and VirB10 from A348, A136(pAB123, pZL48-D5, pZL3) (virB6_Δ_185aa-10, virB1 to virB4) and A136(pAB123, pZL48-D5) (virB6Δ185aa-10). Membrane fractions were separated by sucrose density gradient as described in Materials and Methods. Membrane fractions 3 to 14 (from the top to the bottom of the gradient) were subjected to SDS-PAGE, blotted, and probed with anti-VirB8, anti-VirB9, or anti-VirB10. Similar results were observed in three other experiments. IM, inner membrane; OM, outer membrane.
FIG. 7.
Capacity of VirB9 and VirB10 to form high-molecular-weight complexes in the presence or absence of VirB1 to VirB4. Samples from AS-induced strains were added to loading buffer with DTT (lanes +), added to loading buffer without DTT (lanes −), or cross-linked with BS3 and added to loading buffer with DTT (lanes x). Extracts from equivalent numbers of cells were then resolved by SDS-10% PAGE, blotted, and probed with anti-VirB10 (A) or anti-VirB9 (B) antibodies. Gel 1, A348; gel 2, A136(pAB123, pZL48-D5, pZL3) (virB6_Δ_185aa-10, virB1 to virB4); gel 3, A136(pAB123, pZL48-D5) (virB6_Δ_185aa-10).
Similar articles
- The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid.
Bohne J, Yim A, Binns AN. Bohne J, et al. Proc Natl Acad Sci U S A. 1998 Jun 9;95(12):7057-62. doi: 10.1073/pnas.95.12.7057. Proc Natl Acad Sci U S A. 1998. PMID: 9618538 Free PMC article. - Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion.
Jakubowski SJ, Krishnamoorthy V, Christie PJ. Jakubowski SJ, et al. J Bacteriol. 2003 May;185(9):2867-78. doi: 10.1128/JB.185.9.2867-2878.2003. J Bacteriol. 2003. PMID: 12700266 Free PMC article. - The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus.
Fernandez D, Spudich GM, Zhou XR, Christie PJ. Fernandez D, et al. J Bacteriol. 1996 Jun;178(11):3168-76. doi: 10.1128/jb.178.11.3168-3176.1996. J Bacteriol. 1996. PMID: 8655495 Free PMC article. - Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells.
Zupan JR, Ward D, Zambryski P. Zupan JR, et al. Curr Opin Microbiol. 1998 Dec;1(6):649-55. doi: 10.1016/s1369-5274(98)80110-0. Curr Opin Microbiol. 1998. PMID: 10066547 Review. - Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis.
Kado CI. Kado CI. Mol Microbiol. 1994 Apr;12(1):17-22. doi: 10.1111/j.1365-2958.1994.tb00990.x. Mol Microbiol. 1994. PMID: 7914664 Review.
Cited by
- Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion.
Atmakuri K, Cascales E, Christie PJ. Atmakuri K, et al. Mol Microbiol. 2004 Dec;54(5):1199-211. doi: 10.1111/j.1365-2958.2004.04345.x. Mol Microbiol. 2004. PMID: 15554962 Free PMC article. - Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus.
Judd PK, Kumar RB, Das A. Judd PK, et al. Proc Natl Acad Sci U S A. 2005 Aug 9;102(32):11498-503. doi: 10.1073/pnas.0505290102. Epub 2005 Aug 2. Proc Natl Acad Sci U S A. 2005. PMID: 16076948 Free PMC article. - Analysis of the complete genome sequences of Clostridium perfringens strains harbouring the binary enterotoxin BEC gene and comparative genomics of pCP13-like family plasmids.
Ueda K, Kawahara K, Kimoto N, Yamaguchi Y, Yamada K, Oki H, Yoshida T, Matsuda S, Matsumoto Y, Motooka D, Kawatsu K, Iida T, Nakamura S, Ohkubo T, Yonogi S. Ueda K, et al. BMC Genomics. 2022 Mar 23;23(1):226. doi: 10.1186/s12864-022-08453-4. BMC Genomics. 2022. PMID: 35321661 Free PMC article. - An Agrobacterium VirB10 mutation conferring a type IV secretion system gating defect.
Banta LM, Kerr JE, Cascales E, Giuliano ME, Bailey ME, McKay C, Chandran V, Waksman G, Christie PJ. Banta LM, et al. J Bacteriol. 2011 May;193(10):2566-74. doi: 10.1128/JB.00038-11. Epub 2011 Mar 18. J Bacteriol. 2011. PMID: 21421757 Free PMC article.
References
- Achtman, M., S. Schwuchow, R. Helmuth, G. Morelli, and P. A. Manning. 1978. Cell-cell interactions in conjugating Escherichia coli: Con− mutants and stabilization of mating aggregates. Mol. Gen. Genet. 164:171-183.
- Anthony, K. G., C. Sherburne, R. Sherburne, and L. S. Frost. 1994. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol. Microbiol. 13:939-953. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources