RNA editing and regulation of Drosophila 4f-rnp expression by sas-10 antisense readthrough mRNA transcripts - PubMed (original) (raw)
RNA editing and regulation of Drosophila 4f-rnp expression by sas-10 antisense readthrough mRNA transcripts
Nick T Peters et al. RNA. 2003 Jun.
Abstract
We have previously described an example of extensively A-to-G edited cDNA derived from adult heads of the fruitfly Drosophila melanogaster. In that study, the source of the predicted antisense RNA pairing strand for template recognition by dADAR editase was not identified, and the biological significance of the observed hyperediting was not known. Here, we address each of these questions. 4f-rnp and sas-10 are closely adjacent X-linked genes located on opposite DNA strands that produce convergent transcripts. We show that developmentally regulated antisense sas-10 readthrough mRNA arises by activation of an upstream promoter P2 during the late embryo stage of fly development. The sas-10 readthrough transcripts pair with 4f-rnp mRNA to form double-stranded molecules, as indicated by A-to-G editing observed in both RNA strands. It would be predicted that perfect RNA duplexes would be targeted for modification/degradation by enzyme pathways that recognize double-stranded RNAs, leading to decline in 4f-rnp mRNA levels, and this is what we observe. The observation using quantitative RT-PCR that sas-10 readthrough and 4f-rnp transcript levels are inversely related suggests a role for the antisense RNA in posttranscriptional regulation of 4f-rnp gene expression during development. Potential molecular mechanisms that could lead to this result are discussed, one of which is targeted transcript degradation via the RNAi pathway. Insofar as the dADAR editase and RNAi pathways are known to be constitutive in this system, it is likely that control of antisense RNA transcription is the rate-limiting factor. The results provide insight into roles of naturally occurring antisense RNAs in regulation of eukaryotic gene expression.
Figures
FIGURE 1.
Identification of the fly CG4202 gene as a sas-10 homolog. A BLAST search of the GenBank/EMBL database showed best fit to the yeast and mammalian sas-10 “something-about-silencing” gene. Amino acid sequence alignment shows Drosophila (D.M.) compared to human (H.S.). Shared amino acid identities and conservative changes (+) are shown in center. Putative bipartite nuclear localization signal (NLS) is bracketed. About 40% of the fly amino acid residues are charged.
FIGURE 2.
Quantitative RT-PCR of 4f-rnp and sas-10 transcript levels during development. Orientation diagram shows locations of genes and polarities of their corresponding transcripts. Locations of nested primer sets numbered 1–4 used for PCR are at each end of the arrows. Gene leader (L), trailer (T), and intergenic spacer (S) locations are shown. (A) Representative gels after SYBR Green I staining of cDNA bands. (B) Shorter length sas-10 transcript levels during development, relative to rp49 reference. Numbers of repeat determinations per developmental stage are shown in brackets and standard error bars are indicated. (C) 4f-rnp transcript levels during development, relative to rp49 reference. Labeling is as in (B). (D) Long sas-10 readthrough transcript levels during development, relative to rp49 reference. Labeling is as in (B). Note the roughly 200-fold difference in sas-10 signal intensities between (B) and (D) and also that levels of 4f-rnp and sas-10 readthrough transcripts (C) and (D) are inversely related at late embryo stages of development (15–21 h).
FIGURE 3.
Diagrammatic alignment of compiled genomic sequence organization and sas-10 transcription unit characteristics. Orientation diagrams at top and bottom show relative locations of genes sas-10 and 4f-rnp, separated by 95-bp intergenic spacer (S), with their corresponding leader (L) and conventional trailer (T) tracts. Within poly(A) cDNA clones 4R1#6 (Hunt-Smith 1999) and GH08670 (Drosophila EST database), which both initiate transcription at promoter P1, locations of 5′-termini relative to start codon AUG, putative poly(A) signals (AAUAAA; AAUAUA) relative to 3′-termini, and transcript lengths relative to the start codon (+1) are indicated. Sas-10 transcripts determined by RT-PCR as arising from promoter P2 are labeled with the PCR primers utilized. Their numbering scheme is as for the cDNA clones, and the extent of their final length in vivo is unknown. cDNA clone GH08670 and the shorter RT-PCR transcript depicted overlap with the 4f-rnp trailer to varying extents, while the longer RT-PCR transcript depicted also overlaps the final 251 nt of the 4f-rnp coding region.
FIGURE 4.
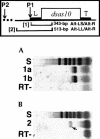
The Drosophila sas-10 gene utilizes dual promoters. Orientation diagram shows locations of gene-specific nested primer sets (brackets) utilized for RT-PCR. In primer set #1 (primers Alt-LS/Alt-R), upstream primer is located just inside 5′-end of cDNA clone 4R1#6, which produces transcripts that terminate at the trailer (T). In primer set #2 (primers Alt-LL/Alt-R), upstream primer is located 170 nt further upstream. (A) Following RT-PCR, gel electrophoresis of products, and cDNA band staining with SYBR Green I, primer set #1 gave intensely staining bands relative to rp49 (Fig. 2A ▶) when using early developmental stage RNA templates including 6–12 h (1a) and 12–18 h (1b). These bands derive from promoter P1, previously known. (B) In contrast, primer set #2 gave a faintly staining band (composition verified by sequencing) relative to rp49 when using later developmental stage RNA templates including 15–21 h embryo (2, short arrow). The faint band is interpreted to derive from a second, distal promoter P2. Densely staining, rapidly migrating bands are due to primer dimers. Sizing standard (S) used during agarose gel electrophoresis was plasmid pBR322 DNA cut with _Alu_I.
FIGURE 5.
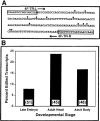
RNA editing in 4f-rnp trailer region and its sas-10 readthrough complement. (A) Compilation of edit site locations (heavy dots). The 23 edits observed are randomly distributed over 12 different sites. (B) Percent edited transcripts from different developmental stages and different locations in the adult fly. Numbers of clones sequenced are indicated for each.
Similar articles
- 5'-UTR mediated translational control of splicing assembly factor RNP-4F expression during development of the Drosophila central nervous system.
Chen J, Yang JT, Doctor DL, Rawlins BA, Shields BC, Vaughn JC. Chen J, et al. Gene. 2013 Oct 10;528(2):154-62. doi: 10.1016/j.gene.2013.07.027. Epub 2013 Jul 25. Gene. 2013. PMID: 23892091 - RNA editing and alternative splicing generate mRNA transcript diversity from the Drosophila 4f-rnp locus.
Petschek JP, Scheckelhoff MR, Mermer MJ, Vaughn JC. Petschek JP, et al. Gene. 1997 Dec 19;204(1-2):267-76. doi: 10.1016/s0378-1119(97)00465-4. Gene. 1997. PMID: 9434193 - RNA editing in Drosophila 4f-rnp gene nuclear transcripts by multiple A-to-G conversions.
Petschek JP, Mermer MJ, Scheckelhoff MR, Simone AA, Vaughn JC. Petschek JP, et al. J Mol Biol. 1996 Jun 28;259(5):885-90. doi: 10.1006/jmbi.1996.0365. J Mol Biol. 1996. PMID: 8683590 - Naturally occurring antisense RNA.
Dolnick BJ. Dolnick BJ. Pharmacol Ther. 1997 Sep;75(3):179-84. doi: 10.1016/s0163-7258(97)00050-8. Pharmacol Ther. 1997. PMID: 9504138 Review. - Naturally occurring antisense RNA: function and mechanisms of action.
Werner A, Sayer JA. Werner A, et al. Curr Opin Nephrol Hypertens. 2009 Jul;18(4):343-9. doi: 10.1097/MNH.0b013e32832cb982. Curr Opin Nephrol Hypertens. 2009. PMID: 19491676 Review.
Cited by
- ADAR RNA editing on antisense RNAs results in apparent U-to-C base changes on overlapping sense transcripts.
Pecori R, Chillón I, Lo Giudice C, Arnold A, Wüst S, Binder M, Marcia M, Picardi E, Papavasiliou FN. Pecori R, et al. Front Cell Dev Biol. 2023 Jan 6;10:1080626. doi: 10.3389/fcell.2022.1080626. eCollection 2022. Front Cell Dev Biol. 2023. PMID: 36684421 Free PMC article. - Long non-coding RNAs: definitions, functions, challenges and recommendations.
Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME, Fitzgerald KA, Gingeras TR, Guttman M, Hirose T, Huarte M, Johnson R, Kanduri C, Kapranov P, Lawrence JB, Lee JT, Mendell JT, Mercer TR, Moore KJ, Nakagawa S, Rinn JL, Spector DL, Ulitsky I, Wan Y, Wilusz JE, Wu M. Mattick JS, et al. Nat Rev Mol Cell Biol. 2023 Jun;24(6):430-447. doi: 10.1038/s41580-022-00566-8. Epub 2023 Jan 3. Nat Rev Mol Cell Biol. 2023. PMID: 36596869 Free PMC article. Review. - Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance.
Li Y, Li X, Yang J, He Y. Li Y, et al. Nat Commun. 2020 Oct 22;11(1):5351. doi: 10.1038/s41467-020-19186-x. Nat Commun. 2020. PMID: 33093449 Free PMC article. - OverGeneDB: a database of 5' end protein coding overlapping genes in human and mouse genomes.
Rosikiewicz W, Suzuki Y, Makalowska I. Rosikiewicz W, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D186-D193. doi: 10.1093/nar/gkx948. Nucleic Acids Res. 2018. PMID: 29069459 Free PMC article. - Novel Stress-Inducible Antisense RNAs of Protein-Coding Loci Are Synthesized by RNA-Dependent RNA Polymerase.
Matsui A, Iida K, Tanaka M, Yamaguchi K, Mizuhashi K, Kim JM, Takahashi S, Kobayashi N, Shigenobu S, Shinozaki K, Seki M. Matsui A, et al. Plant Physiol. 2017 Sep;175(1):457-472. doi: 10.1104/pp.17.00787. Epub 2017 Jul 14. Plant Physiol. 2017. PMID: 28710133 Free PMC article.
References
- Adams, M.D., Rudner, D.Z., and Rio, D.C. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell Biol. 8: 331–339. - PubMed
- Aravin, A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11: 1017–1027. - PubMed
- Arbeitman, M.N., Furlong, E.E.M., Imam, F., Johnson, E., Null, B.H., Baker, B.S., Krasnow, M.A., Scott, M.P., Davis, R.W., and White, K.P. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275. - PubMed
- Ausubel, F., Brent, R., Kingston, R., Moore, D., Seidman, J., Smith, J., and Struhl, K. (eds. F. Ausubel et al.), 1994. –1997. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases