A ligand-binding pocket in the dengue virus envelope glycoprotein - PubMed (original) (raw)
A ligand-binding pocket in the dengue virus envelope glycoprotein
Yorgo Modis et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Dengue virus is an emerging global health threat. Its major envelope glycoprotein, E, mediates viral attachment and entry by membrane fusion. A crystal structure of the soluble ectodomain of E from dengue virus type 2 reveals a hydrophobic pocket lined by residues that influence the pH threshold for fusion. The pocket, which accepts a hydrophobic ligand, opens and closes through a conformational shift in a beta-hairpin at the interface between two domains. These features point to a structural pathway for the fusion-activating transition and suggest a strategy for finding small-molecule inhibitors of dengue and other flaviviruses.
Figures
Fig. 1.
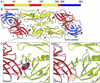
Dengue E protein and its ligand-binding pocket. (A) Domain definition of dengue E. Domain I is red, domain II is yellow, and domain III is blue. (B) The dengue E protein dimer, colored as in A, in complex with β-OG. The β-OG, shown in green, is bound in a hydrophobic pocket under the kl hairpin. A putative receptor-binding loop in domain III (residues 382–385) is marked with a triangle. The glycans in domains I and II are shown in a ball-and-stick representation in red and yellow, respectively. Disulfide bridges are shown in orange. (C) Enlargement of the kl hairpin region, with the structure of dengue E in the absence of β-OG (in gray) superimposed. The strands of the kl hairpin are labeled with “o” or “c” subscripts for the open (β-OG-bound) and closed forms, respectively. The β-OG molecule, shown in a space-filling representation, occupies the ligand-binding pocket. (D) Superposition of the structures of dengue E and TBE E, both in the absence of β-OG. Dengue E is colored as in B, and TBE E is in gray. The view is the same as in C. Figs. 1, 2, and 3 were generated with
bobscript
(33, 34) and
raster 3d
(35).
Fig. 2.
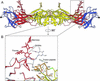
The glycan at residue 153 in dengue 2 virus E protein. (A) The E protein dimer, viewed perpendicular to the dyad axis (and the view in Fig. 1 A). Both glycans are approximately perpendicular to the viral surface. Domain I and the attached glycan are shown in red, domain II and the attached glycan are shown in yellow, and domain III is in blue. Disulfide bridges are shown in orange. The molecule of β-OG bound in the hydrophobic pocket underneath the kl hairpin is in green. A putative receptor-binding loop in domain III (residues 382–385) is marked with a triangle. (B) Enlargement of the area surrounding the glycan at residue 153 in domain I, with the structure of TBE envelope protein superimposed (gray) onto domain I of dengue virus E protein. The fusion peptide is highlighted in orange. The disulfide bridge between residues 92 and 105 is shown in green.
Fig. 3.
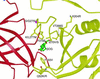
Mutations affecting the pH threshold of fusion (or virulence) in flaviviruses (–41). The mutated residues line the interior of the ligand-binding pocket. For unconserved residues, the residue type in the virus in which the mutation was identified is listed first, followed by the residue type in dengue 2. The coloring is the same as in Fig. 1.
Fig. 4.
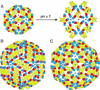
Proposed subunit packing interactions in various flaviviral icosahedral assemblies. (A) Suggested transition from the previously studied T = 1 subviral particles (12) to the fusion-competent T = 1 particle at low pH. On acidification, domain II is proposed to swing out about a hinge at the domain I/II interface, creating homotrimeric contacts at the threefold axis. Clusters of three fusion peptides are displayed at the tip of each trimer. (B) The packing in T = 3 virus-like particles deduced from image reconstructions of dengue virions (13). The 180 subunits are not related by local threefold symmetry. (C) Suggested T = 3 packing intermediate for the virion at low pH (13). E is shown in its native (high pH) conformation. Because all monomers are related by local threefold symmetry, the low-pH conformational change will result in the formation of trimers, as in A.
Comment in
- Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry.
Rey FA. Rey FA. Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):6899-901. doi: 10.1073/pnas.1332695100. Epub 2003 Jun 2. Proc Natl Acad Sci U S A. 2003. PMID: 12782795 Free PMC article. No abstract available.
Similar articles
- Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion.
Nayak V, Dessau M, Kucera K, Anthony K, Ledizet M, Modis Y. Nayak V, et al. J Virol. 2009 May;83(9):4338-44. doi: 10.1128/JVI.02574-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244332 Free PMC article. - Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry.
Rey FA. Rey FA. Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):6899-901. doi: 10.1073/pnas.1332695100. Epub 2003 Jun 2. Proc Natl Acad Sci U S A. 2003. PMID: 12782795 Free PMC article. No abstract available. - Structure of the dengue virus envelope protein after membrane fusion.
Modis Y, Ogata S, Clements D, Harrison SC. Modis Y, et al. Nature. 2004 Jan 22;427(6972):313-9. doi: 10.1038/nature02165. Nature. 2004. PMID: 14737159 - Molecular Aspects of the Dengue Virus Infection Process: A Review.
Zonetti LFC, Coutinho MC, de Araujo AS. Zonetti LFC, et al. Protein Pept Lett. 2018;25(8):712-719. doi: 10.2174/0929866525666180709115506. Protein Pept Lett. 2018. PMID: 29984641 Review. - Flavivirus cell entry and membrane fusion.
Smit JM, Moesker B, Rodenhuis-Zybert I, Wilschut J. Smit JM, et al. Viruses. 2011 Feb;3(2):160-171. doi: 10.3390/v3020160. Epub 2011 Feb 22. Viruses. 2011. PMID: 22049308 Free PMC article. Review.
Cited by
- Pichia pastoris-expressed dengue 2 envelope forms virus-like particles without pre-membrane protein and induces high titer neutralizing antibodies.
Mani S, Tripathi L, Raut R, Tyagi P, Arora U, Barman T, Sood R, Galav A, Wahala W, de Silva A, Swaminathan S, Khanna N. Mani S, et al. PLoS One. 2013 May 23;8(5):e64595. doi: 10.1371/journal.pone.0064595. Print 2013. PLoS One. 2013. PMID: 23717637 Free PMC article. - Atomic view of the histidine environment stabilizing higher-pH conformations of pH-dependent proteins.
Valéry C, Deville-Foillard S, Lefebvre C, Taberner N, Legrand P, Meneau F, Meriadec C, Delvaux C, Bizien T, Kasotakis E, Lopez-Iglesias C, Gall A, Bressanelli S, Le Du MH, Paternostre M, Artzner F. Valéry C, et al. Nat Commun. 2015 Jul 20;6:7771. doi: 10.1038/ncomms8771. Nat Commun. 2015. PMID: 26190377 Free PMC article. - Molecular evolution and epidemiology of four serotypes of dengue virus in Thailand from 1973 to 2007.
Chen SP. Chen SP. Epidemiol Infect. 2013 Feb;141(2):419-24. doi: 10.1017/S0950268812000908. Epub 2012 May 14. Epidemiol Infect. 2013. PMID: 22584140 Free PMC article. - Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity.
Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, Henn MR. Parameswaran P, et al. J Virol. 2012 Aug;86(16):8546-58. doi: 10.1128/JVI.00736-12. Epub 2012 May 30. J Virol. 2012. PMID: 22647702 Free PMC article. - Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection.
Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. Sukupolvi-Petty S, et al. J Virol. 2013 Aug;87(16):8826-42. doi: 10.1128/JVI.01314-13. Epub 2013 Jun 19. J Virol. 2013. PMID: 23785205 Free PMC article.
References
- Gubler, D. J. (2002) Trends Microbiol. 10, 100–103. - PubMed
- Burke, D. S. & Monath, T. P. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), pp. 1043–1125.
- Lindenbach, B. D. & Rice, C. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), pp. 991–1041.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases