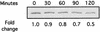pH-regulated gene expression of the gastric pathogen Helicobacter pylori - PubMed (original) (raw)
pH-regulated gene expression of the gastric pathogen Helicobacter pylori
D Scott Merrell et al. Infect Immun. 2003 Jun.
Abstract
Colonization by the gastric pathogen Helicobacter pylori has been shown to be intricately linked to the development of gastritis, ulcers, and gastric malignancy. Little is known about mechanisms employed by the bacterium that help it adapt to the hostile environment of the human stomach. In an effort to extend our knowledge of these mechanisms, we utilized spotted-DNA microarrays to characterize the response of H. pylori to low pH. Expression of approximately 7% of the bacterial genome was reproducibly altered by shift to low pH. Analysis of the differentially expressed genes led to the discovery that acid exposure leads to profound changes in motility of H. pylori, as a larger percentage of acid-exposed bacterial cells displayed motility and moved at significantly higher speeds. In contrast to previous publications, we found that expression of the bacterial virulence gene cagA was strongly repressed by acid exposure. Furthermore, this transcriptional repression was reflected at the level of protein accumulation in the H. pylori cell.
Figures
FIG. 1.
Microarray analysis of the low-pH response of H. pylori. Cluster diagram showing the expression profile of the 118 genes meeting the filter criteria as explained in Materials and Methods after shift to pH 5.0. Two independent experiments are depicted, and relative expression patterns are shown for each time point. Red indicates an increase in expression, while green indicates reduced expression. Representative genes are listed and their relative location indicated by an arrow. A complete list of the regulated factors and the relative changes in expression can be found in Table 1 and as supplementary information at
http://falkow.stanford.edu/whatwedo/supplementarydata/
.
FIG. 2.
The effect of acidic pH on H. pylori motility. A culture of H. pylori was split such that equal portions were suspended at pH 7.0 or 5.0. The response to the different pH conditions was monitored by video microscopy, and the speed of motile bacteria was determined as described in Materials and Methods. Each point represents one motile bacterium at the indicated pH, and the speed of that bacterium is indicated on the y axis. The average speed for each condition is indicated. The statistical significance of the differences in speed between the two sets of pH conditions was determined by using a two-tailed Mann-Whitney test, which revealed a P of 0.0002.
FIG. 3.
The effect of low pH on CagA accumulation. Total protein was harvested from G27 shifted to pH 5.0 brucella broth plus 10% FBS at the times indicated. An equal amount of protein from each point was separated on a 6% polyacrylamide gel, transferred to nitrocellulose membranes, and probed with an anti-CagA specific antibody. The relative change (_n_-fold) from the zero time point is indicated underneath each lane.
FIG. 4.
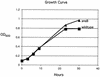
Growth curve analysis of wild-type B128 and its ansB derivative. Brucella broth cultures supplemented with 10% FBS were started from overnight cultures of each strain. Strains were grown in microaerophilic conditions with shaking as described in Materials and Methods, and samples were taken for OD600 measurements at the indicated times.
FIG. 5.
ansB mutants of H. pylori are defective for in vivo competition. Equal concentrations of wild-type and ansB strains were mixed and were used to infect Mongolian gerbils. Colonizing bacteria were recovered after approximately 2.5 to 3 weeks, and the number of CFU of each strain was determined by differential plating. Each circle represents an infected animal, and open circles represent animals for which no ansB derivatives were isolated. Data for two independent experiments are depicted, and the geometric means for each (0.004 and 0.005) are indicated by a black bar.
Similar articles
- Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori.
Gancz H, Censini S, Merrell DS. Gancz H, et al. Infect Immun. 2006 Jan;74(1):602-14. doi: 10.1128/IAI.74.1.602-614.2006. Infect Immun. 2006. PMID: 16369017 Free PMC article. - Expression of cagA, virB/D Complex and/or vacA Genes in Helicobacter pylori Strains Originating from Patients with Gastric Diseases.
Szkaradkiewicz A, Karpiński TM, Linke K, Majewski P, Rożkiewicz D, Goślińska-Kuźniarek O. Szkaradkiewicz A, et al. PLoS One. 2016 Feb 11;11(2):e0148936. doi: 10.1371/journal.pone.0148936. eCollection 2016. PLoS One. 2016. PMID: 26866365 Free PMC article. - Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors.
Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, Wroblewski LE, Piazuelo MB, Correa P, Peek RM Jr. Franco AT, et al. Cancer Res. 2008 Jan 15;68(2):379-87. doi: 10.1158/0008-5472.CAN-07-0824. Cancer Res. 2008. PMID: 18199531 Free PMC article. - Influence of Helicobacter pylori virulence factors CagA and VacA on pathogenesis of gastrointestinal disorders.
Nejati S, Karkhah A, Darvish H, Validi M, Ebrahimpour S, Nouri HR. Nejati S, et al. Microb Pathog. 2018 Apr;117:43-48. doi: 10.1016/j.micpath.2018.02.016. Epub 2018 Feb 9. Microb Pathog. 2018. PMID: 29432909 Review. - The use of microarrays for studying the pathogenesis of Helicobacter pylori.
Morales Espinosa Mdel R, Delgado Sapién G, Cravioto A. Morales Espinosa Mdel R, et al. Rev Latinoam Microbiol. 2003 Jan-Jun;45(1-2):24-9. Rev Latinoam Microbiol. 2003. PMID: 17061518 Review.
Cited by
- The ferric uptake regulator of Helicobacter pylori: a critical player in the battle for iron and colonization of the stomach.
Pich OQ, Merrell DS. Pich OQ, et al. Future Microbiol. 2013 Jun;8(6):725-38. doi: 10.2217/fmb.13.43. Future Microbiol. 2013. PMID: 23701330 Free PMC article. Review. - Analysis of protein expression regulated by the Helicobacter pylori ArsRS two-component signal transduction system.
Loh JT, Gupta SS, Friedman DB, Krezel AM, Cover TL. Loh JT, et al. J Bacteriol. 2010 Apr;192(8):2034-43. doi: 10.1128/JB.01703-08. Epub 2010 Feb 12. J Bacteriol. 2010. PMID: 20154125 Free PMC article. - A new type of intrabacterial nanotransportation system for VacA in Helicobacter pylori.
Wu H, Nakano T, Matsuzaki Y, Ooi Y, Kohno T, Ishihara S, Sano K. Wu H, et al. Med Mol Morphol. 2014 Dec;47(4):224-32. doi: 10.1007/s00795-013-0068-2. Epub 2014 Jan 14. Med Mol Morphol. 2014. PMID: 24420644 - Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods.
Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. Delgado S, et al. Microb Ecol. 2013 Apr;65(3):763-72. doi: 10.1007/s00248-013-0192-5. Epub 2013 Feb 10. Microb Ecol. 2013. PMID: 23397369 - _Helicobacter pylori_l-asparaginase: a study of immunogenicity from an in silico approach.
Belén LH, Beltrán JF, Pessoa A Jr, Castillo RL, de Oliveira Rangel-Yagui C, Farías JG. Belén LH, et al. 3 Biotech. 2022 Nov;12(11):286. doi: 10.1007/s13205-022-03359-0. Epub 2022 Sep 20. 3 Biotech. 2022. PMID: 36276451 Free PMC article.
References
- Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. - PubMed
- Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. - PubMed
- Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. - PubMed
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007502/AI/NIAID NIH HHS/United States
- AI07502-06/AI/NIAID NIH HHS/United States
- R01 CA092229/CA/NCI NIH HHS/United States
- AI38459/AI/NIAID NIH HHS/United States
- CA92229/CA/NCI NIH HHS/United States
- DK56339/DK/NIDDK NIH HHS/United States
- P30 DK056339/DK/NIDDK NIH HHS/United States
- R01 AI038459/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases