A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13 - PubMed (original) (raw)
A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13
Edward M Johnson et al. Nucleic Acids Res. 2003.
Abstract
The MCM8 protein from HeLa cells, a new member of the MCM family, co-isolates through several steps with MCM6 and MCM7, and MCM8 co-immunoprecipitates with MCM4, MCM6 and MCM7, proteins reportedly forming a helicase complex involved in initiation of DNA replication. MCM8 mRNA is expressed in placenta, lung and liver, but is also significantly expressed in adult heart, a tissue with a low percentage of proliferating cells. The MCM8 gene, consisting of 19 exons, is located contrapodal to a gene, consisting of 11 exons, encoding a homolog of the yeast GCD10 gene product. The region between these two transcription units, comprising as few as 62 bp, is TATA-less and highly GC-rich, containing multiple CpG units. MCM8 expression is altered in certain forms of neoplasia. In a case of choriocarcinoma MCM8 mRNA is aberrant, leading to expression of a protein lacking 16 amino acids. In several cases of colon adenocarcinoma MCM8 expression is greatly reduced relative to matched non-cancerous tissue. The potential helicase domain of MCM8 is different from those of other MCM proteins in that it is more homologous to canonical ATP-binding domains of other known helicases. Results suggest that MCM8 may interact with other MCM proteins to alter the function of the replicative MCM protein complex.
Figures
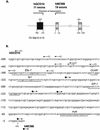
Figure 1
(A) Genomic organization of human MCM8 and GCD10 genes at chromosome band 20p12.3–13. Arrows indicate direction of transcription of the two genes. Gray boxes represent MCM8 exons, black boxes GCD10 exons. Translational start codons are indicated by ATG. MCM8 and GCD10 exon positioning was determined from the genomic sequence, GenBank accession no. AL035461. (B) Genomic DNA sequences in the region of multiple 5′ transcription start points of mRNAs for hMCM8 and hGCD10. Arrowed lines indicate the direction of transcription. Open circles indicate start points for hGCD10, filled circles for hMCM8. Certain potential _cis_-regulatory regions are underlined as indicated. The start point for hMCM8 at +1 is represented in two clones, and that for hGCD10 at –476 is found in three clones. These are at present the most common start points. The closest two start points are separated by 62 bp which are highly GC-rich.
Figure 2
Missing codons in an MCM8 transcript from a case of choriocarcinoma. Genomic DNA in the region of exons 9 and 10 is depicted showing codon triplets. Normal splicing, employing canonical donor and acceptor sites, removes an intron of 242 bp, as shown by the peaked line at the top. Missing codons, representing 16 amino acids, are underlined.
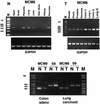
Figure 3
Tissue distribution of expression of MCM8 mRNA. (N, non-cancerous) PCR amplification of an indicator segment comprising 317 bp of MCM8 mRNA or a 269 bp segment comprising the region with a deletion in eight different non-cancerous tissue types. G3PDH housekeeping gene expression is shown as a positive control. PCR amplification is shown at 35 cycles using 1.0 ng first strand cDNA per sample based on reported normalized concentration (Clontech). (T, tumor) PCR amplification of the MCM8 mRNA indicator segment described above in a panel of eight different tumors. G3PDH expression in the tumors is shown as a positive control. PCR products derived from the use of cDNAs from two different colon adenocarcinomas (CX-1 and GI-112) and two different lung carcinomas (LX-1 and GI-117) are included. Total PCR cycles were 34, and 1.0 ng of each first strand cDNA was used for each sample based on reported normalized concentration. (N/T) Expression of MCM8 and ribosomal protein S9 control mRNAs in matched non-cancerous and tumor pairs from the same patient. PCR amplification of the 317 bp MCM8 indicator segment and the 431 bp S9 control using matched human colon adenoid (colon adenocarcinoma) and lung carcinoid (lung malignant carcinoid) non-cancerous and tumor tissue pairs is shown. Specimens were adjusted to 1.0 ng first strand cDNA per sample based on reported initial normalized concentration. PCR amplification is shown at 35 cycles. Left and right markers contained 2 and 4 µg marker DNA, respectively.
Figure 4
Reduced levels of MCM8 mRNA in colon adenocarcinoma. All samples of matched non-cancerous (N) and tumor (T) mRNAs were commercially prepared with first strand cDNA synthesis. PCR was performed as described in Figure 3. Samples for electrophoresis were at 35 and 36 cycles for S9 and MCM8, respectively. A control lane with no template is shown for each set of primers. Densitometry on bands was performed using Photoshop 7.0.
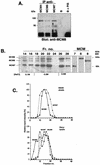
Figure 5
(Next page) Co-isolation and co-immunoprecipitation of MCM8 with other MCM proteins in a potential helicase complex. (A) Co-immunoprecipitation of MCM8 from a HeLa cell lysate using antibodies against either MCM4, MCM6 or MCM7 and agarose beads conjugated to protein G. Antiserum against MCM8 was used in analysis of the blot obtained after gel electrophoresis of the immunoprecipitated samples. Controls are shown to the right: beads without primary antibody (B) and beads with preimmune serum replacing the primary antibody (PIS). (B) Co-isolation of MCM8 with MCM6 and MCM7 after protein purification steps. Overlapping radiographs from western blot analysis following SDS–PAGE indicate the presence of MCM8 (slightly smaller than 100 kDa) co-isolated with MCM6 and MCM7 at various salt concentrations in fractions following DEAE cellulose column chromatography. An immunoblot of the indicated DEAE chromatographic fractions was subjected separately to each of the specific anti-MCM antibodies to obtain three fluorescent radiographic images. These were then precisely superimposed and scanned to obtain the image presented. Individual detection of MCM6, MCM7 and MCM8 in fraction 20 is shown to the right under MCM. (C) Comparison of glycerol gradient centrifugation of MCM8-containing complexes in whole HeLa cell lysate and after elution from a DEAE cellulose column. Glycerol gradients were prepared and subjected to centrifugation and MCM protein levels in fractions quantitated as described in Materials and Methods. The HeLa lysate gradient is in the bottom panel. Multiple peaks for MCM8 (diamonds), two for MCM6 (squares) and one for MCM7 (triangles) can be seen. Fractions from the DEAE column of Figure 4B containing MCM6, MCM7 and MCM8 were pooled and subjected to glycerol gradient centrifugation, shown in the top panel. A single peak for MCM8 (squares) coincides with that for MCM7 (diamonds).
Figure 6
Alignment of MCM protein sequences encompassing potential helicase ATP-binding domains. MCM8 is compared to MCM2–MCM7, proteins implicated in initiation of DNA replication, using the Multalign algorithm (19). Walker A (WA) and Walker B (WB) boxes (14) are underlined in the MCM consensus sequence. A component of the A-box motif, either AKS or SKS, a characteristic of MCM proteins other than MCM8, is indicated with double underlining. In MCM8 this A-box is GKS. Residues conserved for all presented sequences, consensus upper case letters, red in individual sequences. Residues conserved for 50% or more, lower case letters, green. Residues conserved for <50%, white space, black. I or V conserved positions, (!), blue. LM conserved positions, ($), black. NDQE conserved positions, (#), blue.
Similar articles
- Comparative genomics on MCM8 orthologous genes reveals the transcriptional regulation by transcription factor E2F.
Hayashi R, Goto Y, Haga A, Kobayashi D, Ikeda R, Yoshida K. Hayashi R, et al. Gene. 2006 Feb 15;367:126-34. doi: 10.1016/j.gene.2005.10.002. Epub 2005 Dec 1. Gene. 2006. PMID: 16325355 - Colocalization of MCM8 and MCM7 with proteins involved in distinct aspects of DNA replication.
Kinoshita Y, Johnson EM, Gordon RE, Negri-Bell H, Evans MT, Coolbaugh J, Rosario-Peralta Y, Samet J, Slusser E, Birkenbach MP, Daniel DC. Kinoshita Y, et al. Microsc Res Tech. 2008 Apr;71(4):288-97. doi: 10.1002/jemt.20553. Microsc Res Tech. 2008. PMID: 18072282 - Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex.
You Z, Ishimi Y, Masai H, Hanaoka F. You Z, et al. J Biol Chem. 2002 Nov 8;277(45):42471-9. doi: 10.1074/jbc.M205769200. Epub 2002 Aug 30. J Biol Chem. 2002. PMID: 12207017 - MCM family in HCC: MCM6 indicates adverse tumor features and poor outcomes and promotes S/G2 cell cycle progression.
Liu Z, Li J, Chen J, Shan Q, Dai H, Xie H, Zhou L, Xu X, Zheng S. Liu Z, et al. BMC Cancer. 2018 Feb 20;18(1):200. doi: 10.1186/s12885-018-4056-8. BMC Cancer. 2018. PMID: 29463213 Free PMC article. Review. - Archaeal MCM Proteins as an Analog for the Eukaryotic Mcm2-7 Helicase to Reveal Essential Features of Structure and Function.
Miller JM, Enemark EJ. Miller JM, et al. Archaea. 2015 Oct 11;2015:305497. doi: 10.1155/2015/305497. eCollection 2015. Archaea. 2015. PMID: 26539061 Free PMC article. Review.
Cited by
- Function and mechanism of MCM8 in the development and progression of colorectal cancer.
Yu S, Dai W, Zhao S, Yang Y, Xu Y, Wang J, Deng Q, He J, Shi D. Yu S, et al. J Transl Med. 2023 Sep 14;21(1):623. doi: 10.1186/s12967-023-04084-9. J Transl Med. 2023. PMID: 37710286 Free PMC article. - Molecular functions of MCM8 and MCM9 and their associated pathologies.
Helderman NC, Terlouw D, Bonjoch L, Golubicki M, Antelo M, Morreau H, van Wezel T, Castellví-Bel S, Goldberg Y, Nielsen M. Helderman NC, et al. iScience. 2023 Apr 27;26(6):106737. doi: 10.1016/j.isci.2023.106737. eCollection 2023 Jun 16. iScience. 2023. PMID: 37378315 Free PMC article. Review. - High MCM8 expression correlates with unfavorable prognosis and induces immune cell infiltration in hepatocellular carcinoma.
Yu M, Wang H, Xu H, Lv Y, Li Q. Yu M, et al. Aging (Albany NY). 2022 Dec 27;14(24):10027-10049. doi: 10.18632/aging.204440. Epub 2022 Dec 27. Aging (Albany NY). 2022. PMID: 36575045 Free PMC article. - The High Expression of Minichromosome Maintenance Complex Component 5 Is an Adverse Prognostic Factor in Lung Adenocarcinoma.
Sun M, Wang T, Zhu Y, Zhang Y, Zhu L, Li X. Sun M, et al. Biomed Res Int. 2022 Mar 20;2022:4338793. doi: 10.1155/2022/4338793. eCollection 2022. Biomed Res Int. 2022. PMID: 35360518 Free PMC article. - Bioinformatics Analysis Reveals MCM3 as an Important Prognostic Marker in Cervical Cancer.
Ma H, Liu Z, Li H, Guo X, Guo S, Qu P, Wang Y. Ma H, et al. Comput Math Methods Med. 2021 Oct 11;2021:8494260. doi: 10.1155/2021/8494260. eCollection 2021. Comput Math Methods Med. 2021. PMID: 34671420 Free PMC article.
References
- Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. - PubMed
- Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. - PubMed
- Madine M.A., Khoo,C.Y., Mills,A.D. and Laskey,R.A. (1995) MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature, 375, 421–424. - PubMed
- Chong J.P., Mahbubani,H.M., Khoo,C.Y. and Blow,J.J. (1995) Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature, 375, 418–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous