Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America - PubMed (original) (raw)
doi: 10.1086/376435. Epub 2003 May 22.
Michael L Nickerson, Ming-Hui Wei, Michelle B Warren, Gladys M Glenn, Maria L Turner, Laveta Stewart, Paul Duray, Ousman Tourre, Nirmala Sharma, Peter Choyke, Pamela Stratton, Maria Merino, McClellan M Walther, W Marston Linehan, Laura S Schmidt, Berton Zbar
Affiliations
- PMID: 12772087
- PMCID: PMC1180594
- DOI: 10.1086/376435
Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America
Jorge R Toro et al. Am J Hum Genet. 2003 Jul.
Abstract
Hereditary leiomyomatosis and renal cell cancer (HLRCC) is an autosomal dominant disorder characterized by smooth-muscle tumors of the skin and uterus and/or renal cancer. Although the identification of germline mutations in the fumarate hydratase (FH) gene in European families supports it as the susceptibility gene for HLRCC, its role in families in North America has not been studied. We screened for germline mutations in FH in 35 families with cutaneous leiomyomas. Sequence analysis revealed mutations in FH in 31 families (89%). Twenty different mutations in FH were identified, of which 18 were novel. Of these 20 mutations, 2 were insertions, 5 were small deletions that caused frameshifts leading to premature truncation of the protein, and 13 were missense mutations. Eleven unrelated families shared a common mutation: R190H. Eighty-one individuals (47 women and 34 men) had cutaneous leiomyomas. Ninety-eight percent (46/47) of women with cutaneous leiomyomas also had uterine leiomyomas. Eighty-nine percent (41/46) of women with cutaneous and uterine leiomyomas had a total hysterectomy, 44% at age < or =30 years. We identified 13 individuals in 5 families with unilateral and solitary renal tumors. Seven individuals from four families had papillary type II renal cell carcinoma, and another individual from one of these families had collecting duct carcinoma of the kidney. The present study shows that mutations in FH are associated with HLRCC in North America. HLRCC is associated with clinically significant uterine fibroids and aggressive renal tumors. The present study also expands the histologic spectrum of renal tumors and FH mutations associated with HLRCC.
Figures
Figure 1
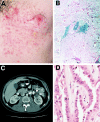
Clinical, histologic, and radiologic manifestations of HLRCC. A, Segmental distribution of leiomyomas in a 32-year-old man. Clinically, cutaneous leiomyomas presented as firm skin-colored to light-brown-colored papules and nodules. B, Histology of cutaneous leiomyoma. Leiomyomas were a proliferation of interlacing bundles of smooth-muscle fibers with a centrally located long blunt-edged nucleus in the dermis. C, Abdominal CT scan, showing the location of a renal tumor in the 32-year-old man shown in panel A. The resected neoplasm was a papillary type II renal cell carcinoma, as shown in panel D. D, Histologic findings of papillary type II renal cell carcinoma. There is a distinct papillary architecture. Cells had an abundant amphophilic cytoplasm and large nuclei with large inclusion-like eosinophilic nucleoli. Magnification × 200.
Figure 2
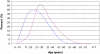
Age distribution at diagnosis of uterine fibroids (red) and reported age distribution at onset of cutaneous leiomyomas (blue)
Figure 3
Physical map of the FH critical region on 1q42.3 A, Ideogram of human chromosome 1q long arm with some markers at 1q42.3-43, with integrated genetic map (above) and physical map (below), showing locations of selected polymorphic markers. Cent. = centromere; Tel. = telomere. B, Expanded physical map. The BAC tiling path is shown by blue horizontal lines and GenBank clone ID numbers. BAC overlaps were confirmed in silico. A sequence gap is present in the 2.6-Mb region. C, Locations of known genes, shown within the 2.6-Mb region of FH. Seven known genes were identified and confirmed by searching public and private databases. D, FH exon-intron structure and mutations identified in the exonic sequence.
Figure 4
FH mutations in patients with HLRCC. Sequencing chromatograms of genomic DNA from control subjects and patients are shown at left (arrows indicate the position of the identified nucleotide changes), and pedigrees are shown at right (asterisks indicate history of renal cancer). The delA at nucleotide 1162 (A), a delG at nucleotide 1339 (D), a delGC at nucleotides 780 and 781 (E), and an 8-bp duplication at nucleotides 1300–1307 (F) cause shifts in the reading frame that lead to a stop codon downstream and truncate the corresponding protein; a missense mutation, G569A (B), changes an arginine to a histidine at codon 190, and another missense mutation, C823T (C), changes a histidine to a tyrosine at codon 275. Sequences containing insertions and deletions are derived from subcloned alleles from affected patients.
Figure 4
FH mutations in patients with HLRCC. Sequencing chromatograms of genomic DNA from control subjects and patients are shown at left (arrows indicate the position of the identified nucleotide changes), and pedigrees are shown at right (asterisks indicate history of renal cancer). The delA at nucleotide 1162 (A), a delG at nucleotide 1339 (D), a delGC at nucleotides 780 and 781 (E), and an 8-bp duplication at nucleotides 1300–1307 (F) cause shifts in the reading frame that lead to a stop codon downstream and truncate the corresponding protein; a missense mutation, G569A (B), changes an arginine to a histidine at codon 190, and another missense mutation, C823T (C), changes a histidine to a tyrosine at codon 275. Sequences containing insertions and deletions are derived from subcloned alleles from affected patients.
Similar articles
- Novel mutations in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer.
Wei MH, Toure O, Glenn GM, Pithukpakorn M, Neckers L, Stolle C, Choyke P, Grubb R, Middelton L, Turner ML, Walther MM, Merino MJ, Zbar B, Linehan WM, Toro JR. Wei MH, et al. J Med Genet. 2006 Jan;43(1):18-27. doi: 10.1136/jmg.2005.033506. Epub 2005 Jun 3. J Med Genet. 2006. PMID: 15937070 Free PMC article. - Association of germline mutations in the fumarate hydratase gene and uterine fibroids in women with hereditary leiomyomatosis and renal cell cancer.
Stewart L, Glenn GM, Stratton P, Goldstein AM, Merino MJ, Tucker MA, Linehan WM, Toro JR. Stewart L, et al. Arch Dermatol. 2008 Dec;144(12):1584-92. doi: 10.1001/archdermatol.2008.517. Arch Dermatol. 2008. PMID: 19075141 Free PMC article. - Hereditary leiomyomatosis and renal cell cancer in families referred for fumarate hydratase germline mutation analysis.
Smit DL, Mensenkamp AR, Badeloe S, Breuning MH, Simon ME, van Spaendonck KY, Aalfs CM, Post JG, Shanley S, Krapels IP, Hoefsloot LH, van Moorselaar RJ, Starink TM, Bayley JP, Frank J, van Steensel MA, Menko FH. Smit DL, et al. Clin Genet. 2011 Jan;79(1):49-59. doi: 10.1111/j.1399-0004.2010.01486.x. Clin Genet. 2011. PMID: 20618355 - Renal cell carcinoma in young FH mutation carriers: case series and review of the literature.
Hol JA, Jongmans MCJ, Littooij AS, de Krijger RR, Kuiper RP, van Harssel JJT, Mensenkamp A, Simons M, Tytgat GAM, van den Heuvel-Eibrink MM, van Grotel M. Hol JA, et al. Fam Cancer. 2020 Jan;19(1):55-63. doi: 10.1007/s10689-019-00155-3. Fam Cancer. 2020. PMID: 31792767 Free PMC article. Review. - Hereditary leiomyomatosis and renal cell cancer (HLRCC).
Kiuru M, Launonen V. Kiuru M, et al. Curr Mol Med. 2004 Dec;4(8):869-75. doi: 10.2174/1566524043359638. Curr Mol Med. 2004. PMID: 15579034 Review.
Cited by
- Preoperative evaluation of renal cell carcinoma by using 18F-FDG PET/CT.
Takahashi M, Kume H, Koyama K, Nakagawa T, Fujimura T, Morikawa T, Fukayama M, Homma Y, Ohtomo K, Momose T. Takahashi M, et al. Clin Nucl Med. 2015 Dec;40(12):936-40. doi: 10.1097/RLU.0000000000000875. Clin Nucl Med. 2015. PMID: 26164183 Free PMC article. - Novel mutations in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer.
Wei MH, Toure O, Glenn GM, Pithukpakorn M, Neckers L, Stolle C, Choyke P, Grubb R, Middelton L, Turner ML, Walther MM, Merino MJ, Zbar B, Linehan WM, Toro JR. Wei MH, et al. J Med Genet. 2006 Jan;43(1):18-27. doi: 10.1136/jmg.2005.033506. Epub 2005 Jun 3. J Med Genet. 2006. PMID: 15937070 Free PMC article. - Multiple/bilateral renal tumors in patients with Birt-Hogg-Dubé syndrome.
Fahmy W, Safwat AS, Bissada NK, Curry N, Guirguis N, Clarke HS, Fraig M, Finkbeiner A. Fahmy W, et al. Int Urol Nephrol. 2007;39(4):995-9. doi: 10.1007/s11255-006-9129-y. Epub 2007 Jan 9. Int Urol Nephrol. 2007. PMID: 17211573 - Fumarate hydratase c.914T > C (p.Phe305Ser) is a pathogenic variant associated with hereditary leiomyomatosis and renal cell cancer syndrome.
Breen KE, Carlo MI, Kemel Y, Maio A, Chen YB, Zhang L, Ceyhan-Birsoy O, Mandelker D. Breen KE, et al. Mol Genet Genomic Med. 2020 Aug;8(8):e1293. doi: 10.1002/mgg3.1293. Epub 2020 May 28. Mol Genet Genomic Med. 2020. PMID: 32463173 Free PMC article. - LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer.
Xie H, Valera VA, Merino MJ, Amato AM, Signoretti S, Linehan WM, Sukhatme VP, Seth P. Xie H, et al. Mol Cancer Ther. 2009 Mar;8(3):626-35. doi: 10.1158/1535-7163.MCT-08-1049. Epub 2009 Mar 10. Mol Cancer Ther. 2009. PMID: 19276158 Free PMC article.
References
Electronic-Database Information
- Celera, http://www.celera.com/
- DHPLC Melt Program, http://insertion.stanford.edu/melt.html
- Ensembl Genome Browser, http://www.ensembl.org/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for mitochondrial FH precursor cDNA [accession number U59309] and BAC genomic sequences [accession numbers AL359764 and AL591898])
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
References
- Akiba T, Hiraga K, Tuboi S (1984) Intracellular distribution of fumarase in various animals. J Biochem 96:189–195 - PubMed
- Alam NA, Bevan S, Churchman M, Barclay E, Barker K, Jaeger EE, Nelson HM, Healy E, Pembroke AC, Friedmann PS, Dalziel K, Calonje E, Anderson J, August PJ, Davies MG, Felix R, Munro CS, Murdoch M, Rendall J, Kennedy S, Leigh IM, Kelsell DP, Tomlinson IP, Houlston RS (2001) Localization of a gene (MCUL1) for multiple cutaneous leiomyomata and uterine fibroids to chromosome 1q42.3-q43. Am J Hum Genet 68:1264–1269 - PMC - PubMed
- Choyke PL, Walther MM, Glenn GM, Wagner JR, Venzon DJ, Lubensky IA, Zbar B, Linehan WM (1997) Imaging features of hereditary papillary renal cancers. J Comput Assist Tomogr 21:737–741 - PubMed
- Coughlin EM, Christensen E, Kunz PL, Krishnamoorthy KS, Walker V, Dennis NR, Chalmers RA, Elpeleg ON, Whelan D, Pollitt RJ, Ramesh V, Mandell R, Shih VE (1998) Molecular analysis and prenatal diagnosis of human fumarase deficiency. Mol Genet Metab 63:254–262 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous