HBXIP functions as a cofactor of survivin in apoptosis suppression - PubMed (original) (raw)
HBXIP functions as a cofactor of survivin in apoptosis suppression
Hiroyuki Marusawa et al. EMBO J. 2003.
Abstract
Survivin is an anti-apoptotic protein that is overexpressed in most human cancers. We show that survivin forms complexes with a cellular protein, hepatitis B X-interacting protein (HBXIP), which was originally recognized for its association with the X protein of hepatitis B virus (HBX). Survivin-HBXIP complexes, but neither survivin nor HBXIP individually, bind pro-caspase-9, preventing its recruitment to Apaf1, and thereby selectively suppressing apoptosis initiated via the mitochondria/cytochrome c pathway. Viral HBX protein also interacts with the survivin- HBXIP complex and suppresses caspase activation in a survivin-dependent manner. Thus, HBXIP functions as a cofactor for survivin, and serves as a link between the cellular apoptosis machinery and a viral pathogen involved in hepatocellular carcinogenesis.
Figures
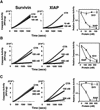
Fig. 1. Differences in caspase suppression by survivin and XIAP. Comparisons were made of the effects of purified survivin (left panels) and XIAP (middle panels) on (A) recombinant active caspase-3 or (B and C) caspase-3-like protease activity induced in cell extracts by Cyt-c. Caspase activity was continuously measured, based on cleavage of Ac-DEVD-AFC fluorigenic substrate [relative fluorescence units (RFU)/min/µg]. In (A), 100 pM recombinant active caspase-3 was incubated with recombinant survivin (left panel) or XIAP (middle panel) or with control protein. In (B), purified survivin (left panel) or XIAP (middle panel), or control protein was added to 293 cell lysates concurrently with Cyt-c and dATP. In (C), survivin (left panel) or XIAP (middle panel) was added to cell extracts after Cyt-c and dATP. Data shown in the panels on the right are normalized relative to control protein (mean ± SD; n = 3).
Fig. 2. HBXIP directly binds survivin. (A) In vitro binding experiments were performed using either purified recombinant survivin (untagged) or GST–XIAP incubated with His6-HBXIP, His6-TRAF3 or SMAC-His6 immobilized on nickel beads. Bound proteins were analyzed by immunoblotting using anti-survivin (upper panel) or anti-XIAP (middle panel) antisera. His6-tagged proteins were also analyzed by SDS–PAGE/immunoblotting using an anti-His6 antibody (lower panel). (B) Various fragments of survivin were produced by in vitro translation with [35S]
l
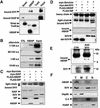
-methionine. Survivin fragments were then incubated with GST–CD40 control protein or GST–HBXIP immobilized on glutathione–Sepharose. Bound proteins were analyzed by autoradiography. The BIR domain is encompassed in residues 15–89. (C and D) 293T cells were transiently transfected with plasmids encoding myc-tagged survivin, mutant survivin lacking the BIR domain (del-SVV) or myc–XIAP, together with FLAG–HBXIP or FLAG–SIP (used as a negative control). Lysates were subjected to immunoprecipitation using anti-FLAG antibody (C and D, upper panel) or anti-myc antibody (D, middle panel). Immunoprecipitates were analyzed by immunoblotting using anti-myc antibody (C and D, upper panel) or anti-FLAG antibody (D, middle panel). Lysates were also blotted by anti-myc (C, middle panel, and D, lower panel) or anti-FLAG antibodies (C, lower panel). (E) Lysates from untransfected HepG2 cells used for immunoprecipitation with rabbit anti-survivin antisera, followed by blotting with chicken anti-HBXIP or rabbit anti-survivin antibodies. (F) Lysates from unfractionated 293 cells (T) and from subcellular fractions (M, membrane; C, cytoplasmic; N, nuclear), normalized for cell equivalents, were analyzed by immunoblotting using anti-HBXIP and anti-survivin. Blotting using anti-Hsp60, anti-caspase-3 and anti-PARP antibodies was also performed as markers for membrane (mitochondria), cytosolic and nuclear proteins, respectively.
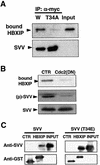
Fig. 3. Interaction of HBXIP and survivin is regulated by phosphorylation. (A) HEK293 cells co-transfected with plasmids encoding FLAG–HBXIP and either wild-type (W) myc–survivin or myc–survivin (T34A) were immunoprecipitated with anti-myc antibody, followed by immunoblotting using anti-FLAG antibody (upper panel). Lysates were also blotted by anti-myc antibody (lower panel). (B) Endogenous survivin was immunoprecipitated from HeLa cells that had been transfected with control or kinase-dead Cdc2 mutant (DN) together with FLAG–HBXIP. Immunoprecipitates were analyzed by immunoblotting using anti-FLAG antibody (upper panel) or an antibody specific for phophorylated Thr34 survivin [(p)-SVV] (middle panel). Lysates were also blotted by anti-survivin antibody (lower panel). (C) In vitro binding experiments were performed, incubating either 0.25 µg of His6-survivin (left panel) or His6-T34E-survivin (right panel) with 1 µg of GST–CD40 (CTR) or GST–HBXIP immobilized on glutathione–Sepharose in 0.5 ml of binding buffer. Bound proteins were analyzed by immunoblotting using anti-survivin (upper panel) antisera. ‘Input’ represents 25 ng of His6-survivin or His6-T34E- survivin. GST-tagged proteins were also analyzed by immunoblotting using anti-GST antibody (lower panel).
Fig. 4. HBXIP collaborates with survivin to suppress Cyt-c-induced caspase activation. Comparisons were made of effects of prior addition of purified HBXIP, survivin or the combination on caspase activity induced in cell extracts by Cyt-c (A), caspase-8 (B) or granzyme B (C), measuring AFC release from Ac-DEVD-AFC. Left panels show representative enzyme progress curves, indicating cumulative release of fluorophore from Ac-DEVD-AFC (RFU/µg). Right panels represent relative enzyme rates (RFU/min/µg) compared with samples treated with control protein (mean ± SD; n = 3 experiments). Several control (CTR) proteins were tested, with representative results shown, here using His6-TRAF3.
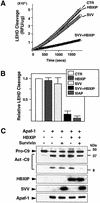
Fig. 5. Combination of HBXIP and survivin inhibits caspase-9 activation. Recombinant pro-caspase-9 was incubated with purified His6-Apaf1, Cyt-c and dATP, with or without recombinant HBXIP, survivin, or the combination of these proteins or various control (CTR) proteins. (A and B) Caspase-9 activity was measured by cleavage of fluorogenic substrate Ac-LEHD-AFC, or (C) proteolytic processing of ∼50 kDa pro-caspase-9 to large (∼35 kDa) and small (∼12 kDa) subunits was analyzed by immunoblotting analysis using anti-caspase-9 antibody. Data in (B) represent mean ± SD (n = 3), while data in (A) show representative enzyme progress curves. In (C), levels of recombinant proteins were also compared by immunoblotting using anti-HBXIP, anti-survivin and anti-His antibodies (lower three panels). Minus signs indicate addition of an equivalent amount of a control protein (GST–CD40 or His6-TRAF3) instead of survivin or HBXIP.
Fig. 6. Combination of HBXIP and survivin inhibits recruitment of pro-caspase-9 to activated Apaf1. (A) GST–survivin, GST–CD40 or GST–HBXIP with or without purified survivin (untagged) was incubated with His6-pro-caspase-9, active His6-caspase-9 (lacking CARD domain) or His6-pro-caspase-3. GST fusion proteins were recovered using glutathione–Sepharose and bound proteins were detected by immunoblotting using anti-caspase-9 or anti-caspase-3 antisera. An equivalent amount of proteins was loaded directly onto gels as a control (‘input’). (B) [35S]pro-caspase-9 was incubated with His6-Apaf1, Cyt-c and dATP, with or without GST–HBXIP and survivin. His6-Apaf1 and associated proteins were recovered by adsorption to Ni-resin, and bound proteins were analyzed by autoradiography (for caspase-9) or immunoblotting using anti-Apaf1 or anti-Cyt-c antibodies. (C) Purified His6-Apaf1, His6-pro-caspase-9, Cyt-c and dATP were incubated without (left panel) or with (right panel) purified survivin and HBXIP. Proteins were analyzed by gel-sieve chromatography using immunoblotting to detect proteins in eluted fractions. (D) Caspase activity in column fractions was determined by monitoring cleavage of Ac-DEVD-AFC after addition of pro-caspase-3.
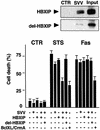
Fig. 7. HBXIP/survivin selectively suppresses caspase-9-dependent apoptosis. Upper panel: full-length HBXIP and HBXIP(1–61) proteins were produced as GST fusions, purified and then tested for binding to His6-survivin or control protein (His6-TRAF3). Bound proteins were analyzed by immunoblotting using anti-GST antisera (upper panel). An equivalent amount of GST fusion protein was loaded directly in gels as a control (‘input’). Lower panel: HT1080 cells were transiently transfected with expression plasmids encoding myc–survivin, FLAG–HBXIP, FLAG–HBXIP(1–61), Bcl-XL (for STS) or CrmA (for anti-Fas), alone or in various combinations, with pEGFP marker plasmid, using pcDNA3 to normalize total DNA content. Then cells were cultured with STS or anti-Fas antibody, and the percentage of apoptotic cells was determined by DAPI staining (mean ± SD; n = 3) a day later.
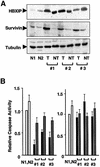
Fig. 8. Elevations of survivin and HBXIP associated with caspase suppression. (A) Protein samples from tumors (T) and non-malignant tissue (NT) of three patients with HBV-related hepatocellular carcinoma and two normal livers specimens (N1 and N2) were normalized for protein content, and analyzed by SDS–PAGE/immunoblotting using antisera specific for HBXIP, survivin or α-tubulin. (B) Caspase activity in the same tissue samples was measured using the fluorigenic substrate Ac-DEVD-AFC after addition of Cyt-c and dATP (left panel) or granzyme B (right panel). Results are expressed relative to caspase activity generated in the normal liver specimen, N1 (mean ± SD; n = 3).
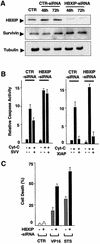
Fig. 9. Regulation of caspase activation and apoptosis by endogenous HBXIP. (A) HeLa cells were transfected with siRNA targeting HBXIP or control RNA (CTR), and cells were analyzed by immunoblotting, using anti-HBXIP (upper panel), anti-survivin (middle) and anti-α- tubulin (lower) antisera, thus verifying siRNA-mediated reductions in endogenous HBXIP expression. (B) Extracts prepared from HeLa cells after treatment with HBXIP–siRNA or control RNA were incubated with Cyt-c and dATP, in the presence or absence of recombinant survivin (left panel) or XIAP (right panel), and caspase activity was measured based on release of AFC from Ac-DEVD-AFC substrate [mean ± SD; n = 3). (C) The percentage of apoptotic cells (mean + SD; n = 3) was determined by DAPI staining following culture of control RNA- or HBXIP siRNA-transfected HeLa cells with 25 µM VP-16 or STS.
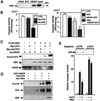
Fig. 10. The X protein of the hepatitis B virus (HBX) associates with survivin through HBXIP and suppresses caspase activation. (A) In vitro protein binding assays were performed using His6-HBX incubated with GST–HBXIP, GST–survivin or GST–CD40 (control) immobilized on glutathione–Sepharose. Bound proteins were analyzed by immunoblotting using anti-HBX antibody. (B) His6-HBX or His6-Traf3 (CTR) proteins, purified His6-HBXIP, purified survivin or various combinations of these proteins were added to 293 cell extracts, normalizing all samples for total protein added using control recombinant proteins. Cyt-c and dATP were then added, and caspase activity was measured 0.5 h later, based on hydrolysis of Ac-DEVD-AFC, and compared with samples treated with control proteins (mean ± SD; n = 3). (C) HEK293 cells were transiently transfected with plasmids encoding FLAG-tagged-HBX or FLAG–SIP (as a control) together with Myc-survivin or HA–HBXIP or control plasmid. Lysates were subjected to immunoprecipitation using anti-FLAG antibody, demonstrating increased association of survivin with HBX when HBXIP was co-expressed (compare the last two lanes on the right). Immunoprecipitates were analyzed by immunoblotting using anti-survivin antibody (upper panel). Lysates were also analyzed by immunoblotting using anti-HBX (middle panel) or anti-HA antibodies (lower panel), confirming protein production. (D) His6-pro-caspase-9 was incubated with GST–HBXIP(+) or GST–CD40 control protein (–), with or without His6-HBX and untagged survivin. GST fusion proteins were recovered on glutathione–Sepharose and bound proteins were detected by immunoblotting using anti-caspase-9, anti-survivin or anti-HBX antisera. (E) HepG2 cell extracts were immunodepleted using anti-survivin antisera or preimmune serum (CTR), and equivalent amounts were analyzed by immunoblotting using anti-survivin antibody (top panel). Then equivalent volumes of extracts were analyzed for caspase activity based on Ac-DEVD-AFC cleavage, where lysates were incubated with HBX (+) or control protein (–) prior to adding Cyt-c and dATP.
Similar articles
- Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP.
Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang Q, Cai N, Wang H, Liu F, Ye L, Zhang X. Zhang W, et al. Mol Cancer. 2014 May 28;13:128. doi: 10.1186/1476-4598-13-128. Mol Cancer. 2014. PMID: 24886421 Free PMC article. - Structural characterization of HBXIP: the protein that interacts with the anti-apoptotic protein survivin and the oncogenic viral protein HBx.
Garcia-Saez I, Lacroix FB, Blot D, Gabel F, Skoufias DA. Garcia-Saez I, et al. J Mol Biol. 2011 Jan 14;405(2):331-40. doi: 10.1016/j.jmb.2010.10.046. Epub 2010 Nov 6. J Mol Biol. 2011. PMID: 21059355 - Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation.
Wen Y, Golubkov VS, Strongin AY, Jiang W, Reed JC. Wen Y, et al. J Biol Chem. 2008 Feb 1;283(5):2793-803. doi: 10.1074/jbc.M708419200. Epub 2007 Nov 21. J Biol Chem. 2008. PMID: 18032378 - Stimulation of cellular proliferation by hepatitis B virus X protein.
Madden CR, Slagle BL. Madden CR, et al. Dis Markers. 2001;17(3):153-7. doi: 10.1155/2001/571254. Dis Markers. 2001. PMID: 11790880 Free PMC article. Review. - An IAP in action: the multiple roles of survivin in differentiation, immunity and malignancy.
Zangemeister-Wittke U, Simon HU. Zangemeister-Wittke U, et al. Cell Cycle. 2004 Sep;3(9):1121-3. Epub 2004 Sep 15. Cell Cycle. 2004. PMID: 15326382 Review.
Cited by
- Modulation of apoptotic signaling by the hepatitis B virus X protein.
Rawat S, Clippinger AJ, Bouchard MJ. Rawat S, et al. Viruses. 2012 Nov 8;4(11):2945-72. doi: 10.3390/v4112945. Viruses. 2012. PMID: 23202511 Free PMC article. Review. - Dynamic protein-protein interaction subnetworks of lung cancer in cases with smoking history.
Yu W, He LR, Zhao YC, Chan MH, Zhang M, He M. Yu W, et al. Chin J Cancer. 2013 Feb;32(2):84-90. doi: 10.5732/cjc.012.10099. Epub 2012 Nov 13. Chin J Cancer. 2013. PMID: 23149315 Free PMC article. - Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection.
Dohi T, Xia F, Altieri DC. Dohi T, et al. Mol Cell. 2007 Jul 6;27(1):17-28. doi: 10.1016/j.molcel.2007.06.004. Mol Cell. 2007. PMID: 17612487 Free PMC article. - Relation between expression pattern of p53 and survivin in cutaneous basal cell carcinomas.
Adamkov M, Halasova E, Rajcani J, Bencat M, Vybohova D, Rybarova S, Galbavy S. Adamkov M, et al. Med Sci Monit. 2011 Feb 25;17(3):BR74-80. doi: 10.12659/msm.881442. Med Sci Monit. 2011. PMID: 21358596 Free PMC article. - Postnatal expansion of the pancreatic beta-cell mass is dependent on survivin.
Jiang Y, Nishimura W, Devor-Henneman D, Kusewitt D, Wang H, Holloway MP, Dohi T, Sabo E, Robinson ML, Altieri DC, Sharma A, Altura RA. Jiang Y, et al. Diabetes. 2008 Oct;57(10):2718-27. doi: 10.2337/db08-0170. Epub 2008 Jul 3. Diabetes. 2008. PMID: 18599523 Free PMC article.
References
- Ambrosini G., Adida,C. and Altieri,D.C. (1997) A novel anti-apoptosis gene, Survivin, expressed in cancer and lymphoma. Nat. Med., 3, 917–921. - PubMed
- Banks D.P., Plescia,J., Altieri,D.C., Chen,J., Rosenberg,S.H., Zhang,H. and Ng,S.C. (2000) Survivin does not inhibit caspase-3 activity. Blood, 96, 4002–4003. - PubMed
- Cryns V. and Yuan,Y. (1998) Proteases to die for. Genes Dev., 12, 1551–1570. - PubMed
- Deveraux Q.L. and Reed,J.C. (1999) IAP family proteins: suppressors of apoptosis. Genes Dev., 13, 239–252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases