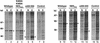Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates - PubMed (original) (raw)
Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates
Christopher R Trotta et al. EMBO J. 2003.
Abstract
60S and 40S ribosomal subunits are assembled in the nucleolus and exported from the nucleus to the cytoplasm independently of each other. We show that in vertebrate cells, transport of both subunits requires the export receptor CRM1 and Ran.GTP. Export of 60S subunits is coupled with that of the nucleo- cytoplasmic shuttling protein NMD3. Human NMD3 (hNMD3) contains a CRM-1-dependent leucine-rich nuclear export signal (NES) and a complex, dispersed nuclear localization signal (NLS), the basic region of which is also required for nucleolar accumulation. When present in Xenopus oocytes, both wild-type and export-defective mutant hNMD3 proteins bind to newly made nuclear 60S pre-export particles at a late step of subunit maturation. The export-defective hNMD3, but not the wild-type protein, inhibits export of 60S subunits from oocyte nuclei. These results indicate that the NES mutant protein competes with endogenous wild-type frog NMD3 for binding to nascent 60S subunits, thereby preventing their export. We propose that NMD3 acts as an adaptor for CRM1-Ran.GTP-mediated 60S subunit export, by a mechanism that is conserved from vertebrates to yeast.
Figures
Fig. 1. NMD3 is conserved between Eukarya and Archaea. (A) Schematic diagram showing the conserved nature of the eukaryotic NMD3 proteins. This alignment was made using MACAW. Blocks of amino acid similarities are indicated by wide boxes, and positions of identity among all four sequences are indicated by vertical bars. (B) Sequence alignment of the C-terminal domains of Homo sapiens (Hs), Drosophila melanogaster (Dm), Saccharomyces cerevisiae (Sc) and Arabidopsis thaliana (At) NMD3 proteins. Conserved (black) and similar (gray) amino acid residues are indicated. NLS and NES indicate the regions resembling a basic nuclear localization signal and a leucine-rich nuclear export signal, respectively. Accession numbers are: Hs (ADD27716), Dm (AE003423.1), At (AAD24816) and Sc (S48909).
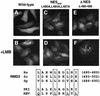
Fig. 2. hNMD3 is a nucleo-cytoplasmic shuttling protein. Wild-type and NES-deficient mutant GFP–hNMD3 proteins were expressed in transiently transfected HeLa cells, and their intracellular distributions were determined by direct fluorescence microscopy after 16 h of expression (A, C and E) and after further treatment of the transfected cells with leptomycin B (3 ng/ml) (+LMB) for 1.5 h (B, D and F). Bottom: comparison of the leucine-rich regions of human and yeast NMD3 proteins with the known NESs of PKI and Rev proteins. Western blot analyses using antibodies to GFP showed that more than ∼95% of the GFP was in the chimeric protein (data not shown), so fluorescence reflects the distribution of the intact fusion protein.
Fig. 3. The NES region of hNMD3 directs Ran·GTP-dependent nuclear export of a heterologous protein in Xenopus oocytes. GST fusion proteins containing the wild-type or NESmut version of the C-terminal 79 amino acids (residues 425–503) of hNMD3 were injected into nuclei of control oocytes (top and middle panels) or oocytes pre-injected with RanT24N (bottom panel) to inhibit Ran·GTP-dependent export, and export was monitored with time. One oocyte equivalent of nuclear (N) and cytoplasmic (C) extracts was analyzed by western blotting with anti-GST antibodies. Note the shorter time course for the top (15, 30 and 60 min) versus the middle and bottom (30, 60 and 120 min) panels. GST alone does not exit the nucleus during this time course (Askjaer et al., 1999; unpublished data).
Fig. 4. The NLS of hNMD3 is complex. GFP–hNMD3 fusion proteins containing the indicated mutations were expressed in HeLa cells as in Figure 2, and protein localizations were monitored without (A–F) or with further treatment with LMB (G–L). The relevant amino acid sequences of the C-terminal domain of hNMD3 are shown below.
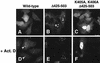
Fig. 5. hNMD3 contains sequences required for nucleolar localization. Wild-type and mutant GFP–hNMD3 proteins were expressed as in Figure 2, and protein localizations were monitored without (A–C) or with treatment with actinomycin D (0.04 ng/ml) for 1.5 h (+ActD) (D–F). Note that treatment with ActD causes the accumulation of GFP–hNMD3 proteins in nucleolar cap structures (arrowheads in D), provided the proteins contain sequences required for nucleolar entry.
Fig. 6. Export of ribosomal subunits in Xenopus oocytes requires CRM1 and Ran·GTP. (A) Outline of the major pathways of rRNA processing in Xenopus (Savino and Gerbi, 1990), including the conversion of 12S rRNA to 6S rRNA, a precursor of 5.8S rRNA, that is matured after export to the cytoplasm (our unpublished results). (B and C) Requirement for the export receptor CRM1. (B) Oocytes were labeled with [32P]GTP at 0 h, and treatment with LMB (400 ng/ml) was initiated at 6 h. The intracellular distributions of newly made rRNAs in control and LMB-treated oocytes were monitored at the indicated times by analysis of 0.5 oocyte equivalents of total nuclear (N) and cytoplasmic (C) RNAs in a 1.2% agarose gel. (C) PKI NES peptides conjugated to BSA (NES–BSA) were injected into nuclei 1 h prior to labeling with [32P]GTP for 24 h, and labeled rRNAs of control and treated oocytes were analyzed as in (A). (D) Requirement for Ran·GTP. Oocytes were labeled with [32P]GTP at 0 h, and RanT24N was injected into the cytoplasm at 24 h; rRNAs of control and treated oocytes were analyzed after 24 and 40 h of labeling, as indicated. The gel mobilities of precursor and mature rRNAs are indicated, and arrowheads show the nuclear accumulation of mature rRNAs upon inhibition of ribosomal subunit export.
Fig. 6. Export of ribosomal subunits in Xenopus oocytes requires CRM1 and Ran·GTP. (A) Outline of the major pathways of rRNA processing in Xenopus (Savino and Gerbi, 1990), including the conversion of 12S rRNA to 6S rRNA, a precursor of 5.8S rRNA, that is matured after export to the cytoplasm (our unpublished results). (B and C) Requirement for the export receptor CRM1. (B) Oocytes were labeled with [32P]GTP at 0 h, and treatment with LMB (400 ng/ml) was initiated at 6 h. The intracellular distributions of newly made rRNAs in control and LMB-treated oocytes were monitored at the indicated times by analysis of 0.5 oocyte equivalents of total nuclear (N) and cytoplasmic (C) RNAs in a 1.2% agarose gel. (C) PKI NES peptides conjugated to BSA (NES–BSA) were injected into nuclei 1 h prior to labeling with [32P]GTP for 24 h, and labeled rRNAs of control and treated oocytes were analyzed as in (A). (D) Requirement for Ran·GTP. Oocytes were labeled with [32P]GTP at 0 h, and RanT24N was injected into the cytoplasm at 24 h; rRNAs of control and treated oocytes were analyzed after 24 and 40 h of labeling, as indicated. The gel mobilities of precursor and mature rRNAs are indicated, and arrowheads show the nuclear accumulation of mature rRNAs upon inhibition of ribosomal subunit export.
Fig. 7. Intracellular distributions of hNMD3 proteins in Xenopus oocytes. Oocytes were injected with m7G-capped mRNAs encoding the indicated wild-type and mutant hNMD3 proteins and labeled with [35S]methionine for 20–24 h; nuclear (N) and cytoplasmic (C) extracts (1 and 0.5 oocyte equivalents, respectively) were analyzed by SDS–PAGE in 8% (lanes 1–8) or 10% (lanes 9–16) gels and by autoradiography. Dots indicate the newly made exogenous hNMD3 proteins. hNMD3(Δ425–503) accumulates to a much lower level in the nucleus than hNMD3(NESmut) (see text), but nonetheless is a very effective inhibitor of 60S subunit export (compare with Figure 8A, lanes 9–12).
Fig. 8. NES-deficient hNMD3 proteins are dominant-negative inhibitors of 60S subunit export. (A) Analyses of large rRNAs. Lanes 1–12: oocytes were pre-labeled with [32P]GTP for ∼6 h prior to injection of m7G-capped mRNAs encoding the indicated hNMD3 proteins, and the intracellular distributions of newly made rRNAs were analyzed after 40–48 h of labeling, as in Figure 6. Lanes 13–18: oocytes were injected with hNMD3 mRNAs 2 h prior to labeling with [32P]GTP, and rRNAs were analyzed after 48 h of labeling. The molar ratios of newly made 28S to18S rRNAs within the nucleus (28S:18S in N) or cytoplasm (28S:18S in C) were determined by quantification of the 32P-labeled rRNAs by phosphorImager analyses; the cytoplasmic ratio of 28S:18S rRNAs is <1.0 in control oocytes due to the slower rate of maturation of 28S rRNA than 18S rRNA (compare with Figure 6B). (B) Analyses of small rRNAs. Lanes 1–12: total RNAs (corresponding to lanes 7–18 in A) were fractionated on 8% denaturing polyacrylamide gels for analyses of 12S and 6S rRNAs (the nuclear precursors of 5.8S rRNA; compare with Figure 6A) and the mature 5S and 5.8S rRNAs. The extra bands seen in the cytoplasm of control oocytes (lane 2) are non-specific degradation products.
Fig. 8. NES-deficient hNMD3 proteins are dominant-negative inhibitors of 60S subunit export. (A) Analyses of large rRNAs. Lanes 1–12: oocytes were pre-labeled with [32P]GTP for ∼6 h prior to injection of m7G-capped mRNAs encoding the indicated hNMD3 proteins, and the intracellular distributions of newly made rRNAs were analyzed after 40–48 h of labeling, as in Figure 6. Lanes 13–18: oocytes were injected with hNMD3 mRNAs 2 h prior to labeling with [32P]GTP, and rRNAs were analyzed after 48 h of labeling. The molar ratios of newly made 28S to18S rRNAs within the nucleus (28S:18S in N) or cytoplasm (28S:18S in C) were determined by quantification of the 32P-labeled rRNAs by phosphorImager analyses; the cytoplasmic ratio of 28S:18S rRNAs is <1.0 in control oocytes due to the slower rate of maturation of 28S rRNA than 18S rRNA (compare with Figure 6B). (B) Analyses of small rRNAs. Lanes 1–12: total RNAs (corresponding to lanes 7–18 in A) were fractionated on 8% denaturing polyacrylamide gels for analyses of 12S and 6S rRNAs (the nuclear precursors of 5.8S rRNA; compare with Figure 6A) and the mature 5S and 5.8S rRNAs. The extra bands seen in the cytoplasm of control oocytes (lane 2) are non-specific degradation products.
Fig. 9. hNMD3 associates with nascent 60S ribosomal subunit in both the nucleus and the cytoplasm. (A) Nucleoplasmic and cytoplasmic extracts of oocytes injected with MBP–hNMD3-NESmut fusion protein and labeled with [32P]GTP were fractionated on 10–40% sucrose gradients, and the distributions of newly made rRNAs (top panels) and MBP–hNMD3 (bottom panels) were determined by agarose gel electrophoresis and western blotting with anti-MBP antibodies, respectively. In the experiment shown, the nucleoplasmic extract was prepared from oocytes treated with VSV M protein (an inhibitor of nuclear export; Her et al., 1997) to increase the levels of 40S and 60S export complexes in the nucleoplasm, but both wild-type and NESmut hNMD3 proteins are also associated with nascent 60S subunits in nucleoplasmic extracts of untreated control oocytes [unpublished data; compare with (B)]. (B and C) Nuclear extracts of oocytes injected with wild-type (WT) or NESmut MBP–hNMD3 proteins were immunoprecipitated with anti-MBP antibodies, and the large (B) and small (C) rRNAs of the bound (Ppt) and unbound (Sup) fractions and the total (Tot) extract were analyzed by gel electrophoresis as in Figure 8. M: marker rRNAs as indicated. Note that the nucleoplasmic extracts are devoid of nucleoli, which contain the majority of the 32S, 36S and 40S precursor rRNA, and which are abundant in total nuclear RNAs (Figure 6B).
Similar articles
- Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway.
Thomas F, Kutay U. Thomas F, et al. J Cell Sci. 2003 Jun 15;116(Pt 12):2409-19. doi: 10.1242/jcs.00464. Epub 2003 Apr 30. J Cell Sci. 2003. PMID: 12724356 - Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex.
West M, Hedges JB, Lo KY, Johnson AW. West M, et al. J Biol Chem. 2007 May 11;282(19):14028-37. doi: 10.1074/jbc.M700256200. Epub 2007 Mar 8. J Biol Chem. 2007. PMID: 17347149 - Nuclear export of ribosomal subunits.
Johnson AW, Lund E, Dahlberg J. Johnson AW, et al. Trends Biochem Sci. 2002 Nov;27(11):580-5. doi: 10.1016/s0968-0004(02)02208-9. Trends Biochem Sci. 2002. PMID: 12417134 Review. - Reengineering ribosome export.
Lo KY, Johnson AW. Lo KY, et al. Mol Biol Cell. 2009 Mar;20(5):1545-54. doi: 10.1091/mbc.e08-10-1000. Epub 2009 Jan 14. Mol Biol Cell. 2009. PMID: 19144820 Free PMC article. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
- Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation.
Bayfield MA, Kaiser TE, Intine RV, Maraia RJ. Bayfield MA, et al. Mol Cell Biol. 2007 May;27(9):3303-12. doi: 10.1128/MCB.00026-07. Epub 2007 Feb 16. Mol Cell Biol. 2007. PMID: 17308035 Free PMC article. - Joining the interface: a site for Nmd3 association on 60S ribosome subunits.
Oeffinger M. Oeffinger M. J Cell Biol. 2010 Jun 28;189(7):1071-3. doi: 10.1083/jcb.201006033. J Cell Biol. 2010. PMID: 20584913 Free PMC article. Review. - Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
Malyutin AG, Musalgaonkar S, Patchett S, Frank J, Johnson AW. Malyutin AG, et al. EMBO J. 2017 Apr 3;36(7):854-868. doi: 10.15252/embj.201696012. Epub 2017 Feb 8. EMBO J. 2017. PMID: 28179369 Free PMC article. - Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells.
Rouquette J, Choesmel V, Gleizes PE. Rouquette J, et al. EMBO J. 2005 Aug 17;24(16):2862-72. doi: 10.1038/sj.emboj.7600752. Epub 2005 Jul 28. EMBO J. 2005. PMID: 16037817 Free PMC article. - Human AATF/Che-1 forms a nucleolar protein complex with NGDN and NOL10 required for 40S ribosomal subunit synthesis.
Bammert L, Jonas S, Ungricht R, Kutay U. Bammert L, et al. Nucleic Acids Res. 2016 Nov 16;44(20):9803-9820. doi: 10.1093/nar/gkw790. Epub 2016 Sep 5. Nucleic Acids Res. 2016. PMID: 27599843 Free PMC article.
References
- Allison L.A., Romaniuk,P.J. and Bakken,A.H. (1991) RNA–protein interactions of stored 5S RNA with TFIIIA and ribosomal protein L5 during Xenopus oogenesis. Dev. Biol., 144, 129–144. - PubMed
- Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous