Molecular basis of p53 functional inactivation by the leukemic protein MLL-ELL - PubMed (original) (raw)
Molecular basis of p53 functional inactivation by the leukemic protein MLL-ELL
Dmitri Wiederschain et al. Mol Cell Biol. 2003 Jun.
Abstract
The Eleven Lysine-rich Leukemia (ELL) gene undergoes translocation and fuses in frame to the Multiple Lineage Leukemia (MLL) gene in a substantial proportion of patients suffering from acute forms of leukemia. Molecular mechanisms of cellular transformation by the MLL-ELL fusion are not well understood. Although both MLL-ELL and wild-type ELL can reduce functional activity of p53 tumor suppressor, our data reveal that MLL-ELL is a much more efficient inhibitor of p53 than is wild-type ELL. We also demonstrate for the first time that ELL extreme C terminus [ELL(eCT)] is required for the recruitment of p53 into MLL-ELL nuclear foci and is both necessary and sufficient for the MLL-ELL inhibition of p53-mediated induction of p21 and apoptosis. Finally, our results demonstrate that MLL-ELL requires the presence of intact ELL(eCT) in order to disrupt p53 interactions with p300/CBP coactivator and thus significantly reduce p53 acetylation in vivo. Since ELL(eCT) has recently been shown to be both necessary and sufficient for MLL-ELL-mediated transformation of normal blood progenitors, our data correlate ELL(eCT) contribution to MLL-ELL transformative effects with its ability to functionally inhibit p53.
Figures
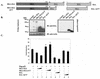
FIG. 1.
ELL(eCT) is necessary and sufficient to inhibit p53 transcriptional activity. (A) Full-length or truncated mutants of MLL-ELL and ELL. (B) The indicated constructs were transfected into 293T cells (5 μg of each cDNA), and cell lysates were prepared 24 h after transfection. Western blot analysis was carried out with the indicated antibodies. Actin served as loading control. Examples of short and long film exposures are shown. (C) Flag-p53 expression vector (100 ng) was cotransfected into H1299 cells with PG13 luciferase reporter (0.25 μg) in the presence or absence of various MLL-ELL and ELL constructs (1 and 2 μg for both) by using the Lipofectamine method. pRL-TK was used as a transfection efficiency control. Both Firefly (PG13) and Renilla (pRL-TK) luciferase activity was measured 36 h posttransfection by using the Promega dual luciferase system. The relative luciferase activity was calculated as a ratio of the Firefly activity to the Renilla activity and is expressed as the fold induction over the vector control (mean ± the standard deviation [SD] of three independent experiments, each done in duplicate). (D) H1299 cells were Lipofectamine transfected as indicated with Flag-p53 (100 ng), Flag-ELL (1 μg, 100 ng, and 10 ng), and MLL-ELL and MLL-ELLΔCT (2 and 4 μg for both). PG13 and pRL-TK plasmids were also included in the transfection mixture. The cells were lysed 36 h posttransfection, and the luciferase activity was determined as described in panel A (the mean ± the SD of three separate experiments, each done in duplicate, is shown in the upper panel). Proteins in the remainder of cell lysates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by Western blot with the indicated antibodies (lower panel). Sample numbers on the luciferase activity chart correspond to those on the Western blot. (E) Cell lysate of 293T cells transfected with Flag-ELL (1 μg) or cell lysates from untransfected cell lines as shown were probed with anti-ELL and anti-p53 antibody. Actin levels were examined to ensure equal loading. N, null; WT, wild type; M, mutant. (F) Schematic depiction of ELL functional domains and ELL truncated mutants. (G) cDNA of ELL or its truncated constructs (5 μg) was transfected into 293T cells. pEGFP (0.5 μg) was included as a transfection efficiency control. Cell lysates were prepared 24 h later and analyzed with the indicated antibody. (H) Plasmids (1 μg) as shown in panel D were cotransfected with Flag-p53 (1 μg), along with PG13-Luc and pRL-TK, into H1299 cells by calcium phosphate precipitation. Luciferase activity, which was recorded 36 h posttransfection, is expressed as the fold induction over vector alone and is shown for three independent transfection experiments, each done in duplicate (mean ± the SD).
FIG. 1.
ELL(eCT) is necessary and sufficient to inhibit p53 transcriptional activity. (A) Full-length or truncated mutants of MLL-ELL and ELL. (B) The indicated constructs were transfected into 293T cells (5 μg of each cDNA), and cell lysates were prepared 24 h after transfection. Western blot analysis was carried out with the indicated antibodies. Actin served as loading control. Examples of short and long film exposures are shown. (C) Flag-p53 expression vector (100 ng) was cotransfected into H1299 cells with PG13 luciferase reporter (0.25 μg) in the presence or absence of various MLL-ELL and ELL constructs (1 and 2 μg for both) by using the Lipofectamine method. pRL-TK was used as a transfection efficiency control. Both Firefly (PG13) and Renilla (pRL-TK) luciferase activity was measured 36 h posttransfection by using the Promega dual luciferase system. The relative luciferase activity was calculated as a ratio of the Firefly activity to the Renilla activity and is expressed as the fold induction over the vector control (mean ± the standard deviation [SD] of three independent experiments, each done in duplicate). (D) H1299 cells were Lipofectamine transfected as indicated with Flag-p53 (100 ng), Flag-ELL (1 μg, 100 ng, and 10 ng), and MLL-ELL and MLL-ELLΔCT (2 and 4 μg for both). PG13 and pRL-TK plasmids were also included in the transfection mixture. The cells were lysed 36 h posttransfection, and the luciferase activity was determined as described in panel A (the mean ± the SD of three separate experiments, each done in duplicate, is shown in the upper panel). Proteins in the remainder of cell lysates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by Western blot with the indicated antibodies (lower panel). Sample numbers on the luciferase activity chart correspond to those on the Western blot. (E) Cell lysate of 293T cells transfected with Flag-ELL (1 μg) or cell lysates from untransfected cell lines as shown were probed with anti-ELL and anti-p53 antibody. Actin levels were examined to ensure equal loading. N, null; WT, wild type; M, mutant. (F) Schematic depiction of ELL functional domains and ELL truncated mutants. (G) cDNA of ELL or its truncated constructs (5 μg) was transfected into 293T cells. pEGFP (0.5 μg) was included as a transfection efficiency control. Cell lysates were prepared 24 h later and analyzed with the indicated antibody. (H) Plasmids (1 μg) as shown in panel D were cotransfected with Flag-p53 (1 μg), along with PG13-Luc and pRL-TK, into H1299 cells by calcium phosphate precipitation. Luciferase activity, which was recorded 36 h posttransfection, is expressed as the fold induction over vector alone and is shown for three independent transfection experiments, each done in duplicate (mean ± the SD).
FIG. 1.
ELL(eCT) is necessary and sufficient to inhibit p53 transcriptional activity. (A) Full-length or truncated mutants of MLL-ELL and ELL. (B) The indicated constructs were transfected into 293T cells (5 μg of each cDNA), and cell lysates were prepared 24 h after transfection. Western blot analysis was carried out with the indicated antibodies. Actin served as loading control. Examples of short and long film exposures are shown. (C) Flag-p53 expression vector (100 ng) was cotransfected into H1299 cells with PG13 luciferase reporter (0.25 μg) in the presence or absence of various MLL-ELL and ELL constructs (1 and 2 μg for both) by using the Lipofectamine method. pRL-TK was used as a transfection efficiency control. Both Firefly (PG13) and Renilla (pRL-TK) luciferase activity was measured 36 h posttransfection by using the Promega dual luciferase system. The relative luciferase activity was calculated as a ratio of the Firefly activity to the Renilla activity and is expressed as the fold induction over the vector control (mean ± the standard deviation [SD] of three independent experiments, each done in duplicate). (D) H1299 cells were Lipofectamine transfected as indicated with Flag-p53 (100 ng), Flag-ELL (1 μg, 100 ng, and 10 ng), and MLL-ELL and MLL-ELLΔCT (2 and 4 μg for both). PG13 and pRL-TK plasmids were also included in the transfection mixture. The cells were lysed 36 h posttransfection, and the luciferase activity was determined as described in panel A (the mean ± the SD of three separate experiments, each done in duplicate, is shown in the upper panel). Proteins in the remainder of cell lysates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by Western blot with the indicated antibodies (lower panel). Sample numbers on the luciferase activity chart correspond to those on the Western blot. (E) Cell lysate of 293T cells transfected with Flag-ELL (1 μg) or cell lysates from untransfected cell lines as shown were probed with anti-ELL and anti-p53 antibody. Actin levels were examined to ensure equal loading. N, null; WT, wild type; M, mutant. (F) Schematic depiction of ELL functional domains and ELL truncated mutants. (G) cDNA of ELL or its truncated constructs (5 μg) was transfected into 293T cells. pEGFP (0.5 μg) was included as a transfection efficiency control. Cell lysates were prepared 24 h later and analyzed with the indicated antibody. (H) Plasmids (1 μg) as shown in panel D were cotransfected with Flag-p53 (1 μg), along with PG13-Luc and pRL-TK, into H1299 cells by calcium phosphate precipitation. Luciferase activity, which was recorded 36 h posttransfection, is expressed as the fold induction over vector alone and is shown for three independent transfection experiments, each done in duplicate (mean ± the SD).
FIG. 1.
ELL(eCT) is necessary and sufficient to inhibit p53 transcriptional activity. (A) Full-length or truncated mutants of MLL-ELL and ELL. (B) The indicated constructs were transfected into 293T cells (5 μg of each cDNA), and cell lysates were prepared 24 h after transfection. Western blot analysis was carried out with the indicated antibodies. Actin served as loading control. Examples of short and long film exposures are shown. (C) Flag-p53 expression vector (100 ng) was cotransfected into H1299 cells with PG13 luciferase reporter (0.25 μg) in the presence or absence of various MLL-ELL and ELL constructs (1 and 2 μg for both) by using the Lipofectamine method. pRL-TK was used as a transfection efficiency control. Both Firefly (PG13) and Renilla (pRL-TK) luciferase activity was measured 36 h posttransfection by using the Promega dual luciferase system. The relative luciferase activity was calculated as a ratio of the Firefly activity to the Renilla activity and is expressed as the fold induction over the vector control (mean ± the standard deviation [SD] of three independent experiments, each done in duplicate). (D) H1299 cells were Lipofectamine transfected as indicated with Flag-p53 (100 ng), Flag-ELL (1 μg, 100 ng, and 10 ng), and MLL-ELL and MLL-ELLΔCT (2 and 4 μg for both). PG13 and pRL-TK plasmids were also included in the transfection mixture. The cells were lysed 36 h posttransfection, and the luciferase activity was determined as described in panel A (the mean ± the SD of three separate experiments, each done in duplicate, is shown in the upper panel). Proteins in the remainder of cell lysates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by Western blot with the indicated antibodies (lower panel). Sample numbers on the luciferase activity chart correspond to those on the Western blot. (E) Cell lysate of 293T cells transfected with Flag-ELL (1 μg) or cell lysates from untransfected cell lines as shown were probed with anti-ELL and anti-p53 antibody. Actin levels were examined to ensure equal loading. N, null; WT, wild type; M, mutant. (F) Schematic depiction of ELL functional domains and ELL truncated mutants. (G) cDNA of ELL or its truncated constructs (5 μg) was transfected into 293T cells. pEGFP (0.5 μg) was included as a transfection efficiency control. Cell lysates were prepared 24 h later and analyzed with the indicated antibody. (H) Plasmids (1 μg) as shown in panel D were cotransfected with Flag-p53 (1 μg), along with PG13-Luc and pRL-TK, into H1299 cells by calcium phosphate precipitation. Luciferase activity, which was recorded 36 h posttransfection, is expressed as the fold induction over vector alone and is shown for three independent transfection experiments, each done in duplicate (mean ± the SD).
FIG. 2.
p53 (aa 1 to 45) is necessary and sufficient to render p53 sensitive to the inhibition by ELL. (A) p53/p73 chimeras were generated by using two-step PCR. p53 deletion mutants were PCR amplified with specific primers. (B) Protein expression of all constructs was verified by a anti-Flag Western blot of 293T-cell lysates transfected with 5 μg of each cDNA. Actin served as an equal loading control. (C) Chimeric or deletion mutants (1 μg) were cotransfected into p53−/− H1299 cells by calcium phosphate precipitation with either Flag-ELL (1 μg) (▪) or with empty vector (□) and with PG13 and pRL-TK plasmids. The relative luciferase activity measured 36 h posttransfection was set to 100% for every construct in the presence of vector alone. All transfections were repeated at least three times, in duplicate experiments (mean ± the SD). (D) Systematic 25-aa deletions in p53(NT) were generated by using standard PCR. (E) Protein expression of p53 deletions was confirmed by a Western blot with anti-Flag antibody. (F) The transcriptional activity of p53 deletion constructs in the presence of vector alone or Flag-ELL was assayed 36 h posttransfection as described in the text. The relative luciferase activity in the presence of vector was set at 100%. All transfections were repeated at least three times, with each done in duplicate (mean ± the SD). (G and H) Protein expression of the indicated p73/p53 chimeric constructs (G) was confirmed by Western blot with anti-Flag (H), and their transcriptional activity was measured by cotransfecting in the presence of either vector alone or Flag-ELL in H1299 cells. (I) The mean ± the SD of three separate transfections, each done in duplicate, is shown.
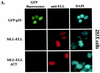
FIG. 3.
Nuclear colocalization of MLL-ELL with p53 requires ELL(eCT) and p53(TAD). 293T (A and B) or U2OS (C and D) cells were cotransfected with 4 μg of the indicated constructs and after 24 h were stained with anti-ELL antibody as described in Materials and Methods. Cells were visualized by using a fluorescence microscope, and representative fields were photographed. The images show GFP-p53 (green); MLL-ELL, ELL, or their truncated mutants (red); and nuclei (DAPI; blue). Merge images were produced by using SPOT advanced software. An enlarged view of representative cells coexpressing various constructs is also shown.
FIG. 3.
Nuclear colocalization of MLL-ELL with p53 requires ELL(eCT) and p53(TAD). 293T (A and B) or U2OS (C and D) cells were cotransfected with 4 μg of the indicated constructs and after 24 h were stained with anti-ELL antibody as described in Materials and Methods. Cells were visualized by using a fluorescence microscope, and representative fields were photographed. The images show GFP-p53 (green); MLL-ELL, ELL, or their truncated mutants (red); and nuclei (DAPI; blue). Merge images were produced by using SPOT advanced software. An enlarged view of representative cells coexpressing various constructs is also shown.
FIG. 3.
Nuclear colocalization of MLL-ELL with p53 requires ELL(eCT) and p53(TAD). 293T (A and B) or U2OS (C and D) cells were cotransfected with 4 μg of the indicated constructs and after 24 h were stained with anti-ELL antibody as described in Materials and Methods. Cells were visualized by using a fluorescence microscope, and representative fields were photographed. The images show GFP-p53 (green); MLL-ELL, ELL, or their truncated mutants (red); and nuclei (DAPI; blue). Merge images were produced by using SPOT advanced software. An enlarged view of representative cells coexpressing various constructs is also shown.
FIG. 3.
Nuclear colocalization of MLL-ELL with p53 requires ELL(eCT) and p53(TAD). 293T (A and B) or U2OS (C and D) cells were cotransfected with 4 μg of the indicated constructs and after 24 h were stained with anti-ELL antibody as described in Materials and Methods. Cells were visualized by using a fluorescence microscope, and representative fields were photographed. The images show GFP-p53 (green); MLL-ELL, ELL, or their truncated mutants (red); and nuclei (DAPI; blue). Merge images were produced by using SPOT advanced software. An enlarged view of representative cells coexpressing various constructs is also shown.
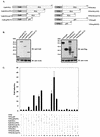
FIG. 4.
In vivo binding between p53 and ELL is mediated by ELL(eCT) and p53(NT). (A) Various segments of p53 and ELL were fused in frame to either Gal4DBD or VP16TAD. (B) Protein expression of all constructs was confirmed by Western blot analysis of 293T-cell lysates with the indicated antibody. (C) Gal4 fusions of ELL were cotransfected into H1299 cells with VP16 constructs of p53 along with pGL3-G5SV and pRL-TK, and luciferase expression was measured as previously described 36 h posttransfection. In a separate experiment, Gal4-p53(CT) was cotransfected into H1299 cells with VP16 ELL fusions as shown. Solid bars represent the fold induction of luciferase activity compared to the vector control (mean ± the SD of three separate transfections, each done in duplicate).
FIG. 5.
MLL-ELL and ELL disrupt p53 interactions with p300 transcriptional coactivator. (A) H1299 cells were transfected with the following constructs as indicated: Flag-p53 (0.3 μg), MLL-ELL (2, 5, and 10 μg), MLL-ELLΔCT (2 and 5 μg), and HA-p300 (1 μg). pEGFP (0.5 μg) served as a transfection efficiency control. Empty vector was used to correct for total DNA amount. Cell lysates were prepared 24 h posttransfection and Western blot analysis was carried out with the indicated antibody. (B) A similar experiment was carried out with 1, 2.5, and 5 μg of ELL and with 1 and 2.5 μg of ELLΔeCT. (C) Flag-p53 plasmid (25 ng) was cotransfected into H1299 cells with PG13 luciferase reporter and with or without Flag-ELL (250 ng) and HA-p300 (2 and 4 μg) as indicated. pRL-TK was included to control for transfection efficiency. Values shown are the fold induction in luciferase expression over the vector control (mean ± the SD of three separate experiments). (D) Gal4 and VP16 fusions of p300 and p53. (E) Protein expression of constructs depicted in panel D was analyzed by transfection of 293T cells, followed by Western blot with anti-Gal4 as described. (F) The indicated plasmids, as well as pGL3-G5SV and pRL-TK plasmids, were cotransfected into H1299 cells in the presence or absence of 2 μg of Flag-ELL or Flag-ELLΔeCT. The luciferase activity measured 36 h posttransfection is expressed as the fold induction over the vector control (mean ± the SD of three independent experiments). (G and H) The indicated Gal4 fusions (G), the protein levels of which were examined by Western blot (H), were transfected into H1299 cells with or without Flag-ELL or Flag-ELLΔeCT and luciferase reporters (pGL3-G5SV and pRL-TK). (I) The luciferase expression was measured as described 36 h after transfection and is shown as the fold induction compared with vector control in three separate transfection experiments (mean ± the SD). (J) The indicated p300 Gal4 and ELL VP16 constructs were cotransfected with pGL3-G5SV and pRL-TK into H1299 cells. (K) The luciferase activity was assessed 36 h posttransfection (the fold induction over the vector control is shown as the mean ± the SD of three separate experiments, each done in duplicate).
FIG. 5.
MLL-ELL and ELL disrupt p53 interactions with p300 transcriptional coactivator. (A) H1299 cells were transfected with the following constructs as indicated: Flag-p53 (0.3 μg), MLL-ELL (2, 5, and 10 μg), MLL-ELLΔCT (2 and 5 μg), and HA-p300 (1 μg). pEGFP (0.5 μg) served as a transfection efficiency control. Empty vector was used to correct for total DNA amount. Cell lysates were prepared 24 h posttransfection and Western blot analysis was carried out with the indicated antibody. (B) A similar experiment was carried out with 1, 2.5, and 5 μg of ELL and with 1 and 2.5 μg of ELLΔeCT. (C) Flag-p53 plasmid (25 ng) was cotransfected into H1299 cells with PG13 luciferase reporter and with or without Flag-ELL (250 ng) and HA-p300 (2 and 4 μg) as indicated. pRL-TK was included to control for transfection efficiency. Values shown are the fold induction in luciferase expression over the vector control (mean ± the SD of three separate experiments). (D) Gal4 and VP16 fusions of p300 and p53. (E) Protein expression of constructs depicted in panel D was analyzed by transfection of 293T cells, followed by Western blot with anti-Gal4 as described. (F) The indicated plasmids, as well as pGL3-G5SV and pRL-TK plasmids, were cotransfected into H1299 cells in the presence or absence of 2 μg of Flag-ELL or Flag-ELLΔeCT. The luciferase activity measured 36 h posttransfection is expressed as the fold induction over the vector control (mean ± the SD of three independent experiments). (G and H) The indicated Gal4 fusions (G), the protein levels of which were examined by Western blot (H), were transfected into H1299 cells with or without Flag-ELL or Flag-ELLΔeCT and luciferase reporters (pGL3-G5SV and pRL-TK). (I) The luciferase expression was measured as described 36 h after transfection and is shown as the fold induction compared with vector control in three separate transfection experiments (mean ± the SD). (J) The indicated p300 Gal4 and ELL VP16 constructs were cotransfected with pGL3-G5SV and pRL-TK into H1299 cells. (K) The luciferase activity was assessed 36 h posttransfection (the fold induction over the vector control is shown as the mean ± the SD of three separate experiments, each done in duplicate).
FIG. 5.
MLL-ELL and ELL disrupt p53 interactions with p300 transcriptional coactivator. (A) H1299 cells were transfected with the following constructs as indicated: Flag-p53 (0.3 μg), MLL-ELL (2, 5, and 10 μg), MLL-ELLΔCT (2 and 5 μg), and HA-p300 (1 μg). pEGFP (0.5 μg) served as a transfection efficiency control. Empty vector was used to correct for total DNA amount. Cell lysates were prepared 24 h posttransfection and Western blot analysis was carried out with the indicated antibody. (B) A similar experiment was carried out with 1, 2.5, and 5 μg of ELL and with 1 and 2.5 μg of ELLΔeCT. (C) Flag-p53 plasmid (25 ng) was cotransfected into H1299 cells with PG13 luciferase reporter and with or without Flag-ELL (250 ng) and HA-p300 (2 and 4 μg) as indicated. pRL-TK was included to control for transfection efficiency. Values shown are the fold induction in luciferase expression over the vector control (mean ± the SD of three separate experiments). (D) Gal4 and VP16 fusions of p300 and p53. (E) Protein expression of constructs depicted in panel D was analyzed by transfection of 293T cells, followed by Western blot with anti-Gal4 as described. (F) The indicated plasmids, as well as pGL3-G5SV and pRL-TK plasmids, were cotransfected into H1299 cells in the presence or absence of 2 μg of Flag-ELL or Flag-ELLΔeCT. The luciferase activity measured 36 h posttransfection is expressed as the fold induction over the vector control (mean ± the SD of three independent experiments). (G and H) The indicated Gal4 fusions (G), the protein levels of which were examined by Western blot (H), were transfected into H1299 cells with or without Flag-ELL or Flag-ELLΔeCT and luciferase reporters (pGL3-G5SV and pRL-TK). (I) The luciferase expression was measured as described 36 h after transfection and is shown as the fold induction compared with vector control in three separate transfection experiments (mean ± the SD). (J) The indicated p300 Gal4 and ELL VP16 constructs were cotransfected with pGL3-G5SV and pRL-TK into H1299 cells. (K) The luciferase activity was assessed 36 h posttransfection (the fold induction over the vector control is shown as the mean ± the SD of three separate experiments, each done in duplicate).
FIG. 5.
MLL-ELL and ELL disrupt p53 interactions with p300 transcriptional coactivator. (A) H1299 cells were transfected with the following constructs as indicated: Flag-p53 (0.3 μg), MLL-ELL (2, 5, and 10 μg), MLL-ELLΔCT (2 and 5 μg), and HA-p300 (1 μg). pEGFP (0.5 μg) served as a transfection efficiency control. Empty vector was used to correct for total DNA amount. Cell lysates were prepared 24 h posttransfection and Western blot analysis was carried out with the indicated antibody. (B) A similar experiment was carried out with 1, 2.5, and 5 μg of ELL and with 1 and 2.5 μg of ELLΔeCT. (C) Flag-p53 plasmid (25 ng) was cotransfected into H1299 cells with PG13 luciferase reporter and with or without Flag-ELL (250 ng) and HA-p300 (2 and 4 μg) as indicated. pRL-TK was included to control for transfection efficiency. Values shown are the fold induction in luciferase expression over the vector control (mean ± the SD of three separate experiments). (D) Gal4 and VP16 fusions of p300 and p53. (E) Protein expression of constructs depicted in panel D was analyzed by transfection of 293T cells, followed by Western blot with anti-Gal4 as described. (F) The indicated plasmids, as well as pGL3-G5SV and pRL-TK plasmids, were cotransfected into H1299 cells in the presence or absence of 2 μg of Flag-ELL or Flag-ELLΔeCT. The luciferase activity measured 36 h posttransfection is expressed as the fold induction over the vector control (mean ± the SD of three independent experiments). (G and H) The indicated Gal4 fusions (G), the protein levels of which were examined by Western blot (H), were transfected into H1299 cells with or without Flag-ELL or Flag-ELLΔeCT and luciferase reporters (pGL3-G5SV and pRL-TK). (I) The luciferase expression was measured as described 36 h after transfection and is shown as the fold induction compared with vector control in three separate transfection experiments (mean ± the SD). (J) The indicated p300 Gal4 and ELL VP16 constructs were cotransfected with pGL3-G5SV and pRL-TK into H1299 cells. (K) The luciferase activity was assessed 36 h posttransfection (the fold induction over the vector control is shown as the mean ± the SD of three separate experiments, each done in duplicate).
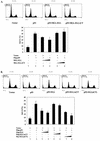
FIG. 6.
ELL(eCT) is necessary and sufficient to suppress both p53- and p53(6KR)-mediated induction of p21. (A) H1299 cells were transfected with the following constructs using Lipofectamine: Flag-p53 (1 μg) or MLL-ELL or MLL-ELLΔCT (5 or 10 μg of each cDNA). pEGFP (0.5 μg) was included as a transfection efficiency control. (B) A similar experiment was conducted with 2 and 5 μg of Flag-ELL, Flag-ELLΔeCT, and Myc-ELL(eCT). Cell lysates prepared 24 h posttransfection were probed with the indicated antibody. (C) H1299 cells transfected with Flag-p53 (0.3 μg), Flag-p53(6KR) (0.3 μg), and HA-p300 (1 μg) were harvested 24 h posttransfection, and cell lysates were analyzed by Western blot with the indicated antibody. (D) H1299 cells were transfected as shown with p53(6KR) (1 μg) and ELL constructs (2 and 5 μg), and after 24 h the cell lysates were analyzed with the indicated antibody.
FIG. 7.
ELL(eCT) is necessary and sufficient to inhibit p53-induced apoptosis. (A) Flag-p53 (1 μg), MLL-ELL or MLL-ELLΔCT (5 and 10 μg each), and Us9-GFP (0.5 μg) were transfected into H1299 cells with Lipofectamine. After 36 h of incubation, cells were fixed in 70% ethanol for 24 to 72 h and stained with propidium iodide solution. Approximately 10,000 GFP-positive cells from each sample were then analyzed for their DNA content by using flow cytometry. The percentage of cells in sub-G1 phase is shown as the mean ± the SD of three independent experiments. Representative histograms from each transfection are also shown. (B) An experiment identical to that in panel A was conducted with Flag-ELL, Flag-ELLΔeCT, and Myc-ELL(eCT) (5 and 10 μg each).
Similar articles
- Multiple mixed lineage leukemia (MLL) fusion proteins suppress p53-mediated response to DNA damage.
Wiederschain D, Kawai H, Shilatifard A, Yuan ZM. Wiederschain D, et al. J Biol Chem. 2005 Jul 1;280(26):24315-21. doi: 10.1074/jbc.M412237200. Epub 2005 Apr 25. J Biol Chem. 2005. PMID: 15851483 - A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL.
DiMartino JF, Miller T, Ayton PM, Landewe T, Hess JL, Cleary ML, Shilatifard A. DiMartino JF, et al. Blood. 2000 Dec 1;96(12):3887-93. Blood. 2000. PMID: 11090074 - Regulation of the p53 homolog p73 by adenoviral oncogene E1A.
Das S, El-Deiry WS, Somasundaram K. Das S, et al. J Biol Chem. 2003 May 16;278(20):18313-20. doi: 10.1074/jbc.M211704200. Epub 2003 Mar 13. J Biol Chem. 2003. PMID: 12639967 - Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins.
Ayton PM, Cleary ML. Ayton PM, et al. Oncogene. 2001 Sep 10;20(40):5695-707. doi: 10.1038/sj.onc.1204639. Oncogene. 2001. PMID: 11607819 Review. - Molecular mechanisms of MLL-associated leukemia.
Yokoyama A. Yokoyama A. Int J Hematol. 2015 Apr;101(4):352-61. doi: 10.1007/s12185-015-1774-4. Epub 2015 Mar 17. Int J Hematol. 2015. PMID: 25773519 Review.
Cited by
- Mutational analysis of an RNA polymerase II elongation factor in Drosophila melanogaster.
Gerber MA, Shilatifard A, Eissenberg JC. Gerber MA, et al. Mol Cell Biol. 2005 Sep;25(17):7803-11. doi: 10.1128/MCB.25.17.7803-7811.2005. Mol Cell Biol. 2005. PMID: 16107725 Free PMC article. - Flt3-ITD alters chemotherapy response in vitro and in vivo in a p53-dependent manner.
Pardee TS, Zuber J, Lowe SW. Pardee TS, et al. Exp Hematol. 2011 Apr;39(4):473-485.e4. doi: 10.1016/j.exphem.2011.01.009. Epub 2011 Feb 1. Exp Hematol. 2011. PMID: 21288478 Free PMC article. - The molecular biology of mixed lineage leukemia.
Slany RK. Slany RK. Haematologica. 2009 Jul;94(7):984-93. doi: 10.3324/haematol.2008.002436. Epub 2009 Jun 16. Haematologica. 2009. PMID: 19535349 Free PMC article. Review. - Atg5-dependent autophagy contributes to the development of acute myeloid leukemia in an MLL-AF9-driven mouse model.
Liu Q, Chen L, Atkinson JM, Claxton DF, Wang HG. Liu Q, et al. Cell Death Dis. 2016 Sep 8;7(9):e2361. doi: 10.1038/cddis.2016.264. Cell Death Dis. 2016. PMID: 27607576 Free PMC article. - Elongation factor ELL (Eleven-Nineteen Lysine-rich Leukemia) acts as a transcription factor for direct thrombospondin-1 regulation.
Zhou J, Feng X, Ban B, Liu J, Wang Z, Xiao W. Zhou J, et al. J Biol Chem. 2009 Jul 10;284(28):19142-52. doi: 10.1074/jbc.M109.010439. Epub 2009 May 15. J Biol Chem. 2009. PMID: 19447890 Free PMC article.
References
- Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. - PubMed
- Butler, L. H., R. Slany, X. Cui, M. L. Cleary, and D. Y. Mason. 1997. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood 89:3361-3370. - PubMed
- DiMartino, J. F., T. Miller, P. M. Ayton, T. Landewe, J. L. Hess, M. L. Cleary, and A. Shilatifard. 2000. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood 96:3887-3893. - PubMed
- Downes, M., L. J. Burke, P. J. Bailey, and G. E. Muscat. 1996. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with Rev-erbA alpha and RVR: physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res. 24:4379-4386. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous