The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin - PubMed (original) (raw)
The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin
David R Kovar et al. J Cell Biol. 2003.
Abstract
Cytokinesis in most eukaryotes requires the assembly and contraction of a ring of actin filaments and myosin II. The fission yeast Schizosaccharomyces pombe requires the formin Cdc12p and profilin (Cdc3p) early in the assembly of the contractile ring. The proline-rich formin homology (FH) 1 domain binds profilin, and the FH2 domain binds actin. Expression of a construct consisting of the Cdc12 FH1 and FH2 domains complements a conditional mutant of Cdc12 at the restrictive temperature, but arrests cells at the permissive temperature. Cells overexpressing Cdc12(FH1FH2)p stop growing with excessive actin cables but no contractile rings. Like capping protein, purified Cdc12(FH1FH2)p caps the barbed end of actin filaments, preventing subunit addition and dissociation, inhibits end to end annealing of filaments, and nucleates filaments that grow exclusively from their pointed ends. The maximum yield is one filament pointed end per six formin polypeptides. Profilins that bind both actin and poly-l-proline inhibit nucleation by Cdc12(FH1FH2)p, but polymerization of monomeric actin is faster, because the filaments grow from their barbed ends at the same rate as uncapped filaments. On the other hand, Cdc12(FH1FH2)p blocks annealing even in the presence of profilin. Thus, formins are profilin-gated barbed end capping proteins with the ability to initiate actin filaments from actin monomers bound to profilin. These properties explain why contractile ring assembly requires both formin and profilin and why viability depends on the ability of profilin to bind both actin and poly-l-proline.
Figures
Figure 1.
Fission yeast formin Cdc12 domains, purified protein, complementation of a temperature-sensitive cdc12 mutant, and overexpression of Cdc12(FH1FH2)p in wild-type cells. (A) Domain organization of Cdc12p and the Cdc12(FH1FH2)p construct. (B) Purified proteins separated by SDS-PAGE and stained with Coomassie blue; (lane 1) Cdc12(FH1FH2)p, (lane 2) mouse capping protein _Mm_CP, (lane 3) wild-type fission yeast profilin _Sp_PRF, (lane 4) profilin mutant _Sp_PRF-K81E, (lane 5) profilin mutant _Sp_PRF-Y5D. Molecular weights are indicated on the left. (C) The formin fragment Cdc12(FH1FH2)p complements a temperature-sensitive cdc12 mutant (cdc12-112). Cdc12-112 cells with either a control (pREP41) or a pREP41-cdc12(FH1FH2) plasmid were grown in the absence of thiamine to induce expression of Cdc12(FH1FH2)p. Overexpression of Cdc12(FH1FH2)p allowed growth of cdc12-112 cells at the restrictive temperature (36°C) but arrested cells at the permissive temperature (25°C) on minimal media (EMM) plates after 72 h. (D–H) Overexpression of Cdc12(FH1FH2)p in wild-type cells. Wild-type cells with either a control (pREP41) or a pREP41-cdc12(FH1FH2) plasmid were grown in the absence of thiamine to induce expression of Cdc12(FH1FH2)p. (D) Differential interference contrast (DIC) and fluorescence micrographs of control cells and cells overexpressing Cdc12(FH1FH2)p for 24 h at 25°C in minimal (EMM) liquid media. Cells were stained for nuclei and septa (Hoechst) or filamentous actin (rhodamine-phalloidin). Cells overexpressing Cdc12(FH1FH2)p had multiple nuclei, partial and misoriented septa, and increased actin aggregates and cables, but lack actin patches and contractile rings. Bar, 5 μm. (E–H) Quantitation of the time course of morphological features of cells with a (□) pREP41 control plasmid or a (○) pREP41-cdc12(FH1FH2) plasmid overexpressing Cdc12(FH1FH2)p by removal of thiamine at time zero. (E) Cell density (OD595). (F) Percent of cells with multiple nuclei. (G) Percent of cells with normal septa and (•) abnormal septa for pREP41-cdc12(FH1FH2). (H) Percent of cells with contractile rings marked with a myosin essential light chain (GFP–Cdc4p).
Figure 2.
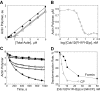
Effect of Cdc12(FH1FH2)p and mouse capping protein ( Mm CP) on actin depolymerization and critical concentration. The conditions were as follows: 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 0.2 mM ATP, 90 μM CaCl2, and 0.25% glycerol at 25°C. (A and B) Critical concentration for assembly of rabbit skeletal muscle actin. (A) Dependence of actin polymer concentration on total actin concentration in the (•) absence or presence of either (□) 10 nM _Mm_CP or (○) 100 nM Cdc12(FH1FH2)p. Actin (5% pyrene labeled) was polymerized for 16 h. The polymer concentration was measured from the pyrene fluorescence, plotted versus actin concentration, and fit by linear regression. The C c values were 0.1 μM for actin alone, 0.9 μM with 100 nM Cdc12(FH1FH2)p, and 1.0 μM with 10 nM _Mm_CP. (B) Dependence of the polymer concentration of 1 μM actin on the concentration of Cdc12(FH1FH2)p. 5 μM actin filaments (10% pyrene labeled) were diluted to 1 μM in the presence of a range of Cdc12(FH1FH2)p concentrations. After 16 h, the pyrene fluorescence was measured, and the actin polymer concentration was plotted versus the log of Cdc12(FH1FH2)p concentration. (C and D) Time course of depolymerization of 5 μM actin filaments (70% pyrene labeled) after dilution to 0.1 μM in the presence of a range of concentrations of Cdc12(FH1FH2)p, (•) 0, (▵) 7.5 nM, and (▴) 25 nM, or a range of concentrations of _Mm_CP, (•) 0, (□) 2.5 nM, and (▪) 10 nM. (D) Dependence of the rate of depolymerization on the concentration of (•) Cdc12(FH1FH2)p or (○) _Mm_CP. The data from 300–1,000 s of each curve was fit with single exponentials, and the depolymerization rates were expressed as a fraction of the rate of actin alone.
Figure 3.
Real-time visualization of actin assembly in the presence of Cdc12(FH1FH2)p or mouse capping protein by TIRF microscopy. The conditions were as follows: 3 μM (25% Oregon green 488–labeled) Mg-ATP actin, 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 0.5% methylcellulose at 25°C. Flow cells were coated with NEM-myosin to capture the actin filaments. Direct comparisons of samples stained with rhodamine-phalloidin and adsorbed to coverslips coated with poly-
l
-lysine (Fig. 4) showed that the conditions of these TIRF experiments strongly favored the observation of long filaments. In fact, we observed no filaments at concentrations of Cdc12(FH1FH2)p >500 nM, where the biochemical results (and rhodamine-phalloidin staining) indicated numerous short filaments. (A–C) Images of actin filaments 500 s after initiating the reactions. Bar, 10 μm or 3,700 subunits. (A) Actin alone. (B) Actin with 50 nM _Mm_CP. Both long filaments (n = 15) and short filaments (n = 12; arrowheads) grew at a pointed end rate of 0.5 ± 0.1 subunits/s. (C) Actin with 250 nM Cdc12(FH1FH2)p. (D–F) Measurements of changes in filament length with time. Control filament barbed ends grew at 19.0 ± 0.1 subunits/s (n = 16 filaments), and pointed ends grew at 0.8 ± 0.1 subunits/s (n = 16). (D) Barbed ends in the presence of Cdc12(FH1FH2)p. The longer filaments (dashed line) grew at 25.2 ± 0.2 subunits/s (n = 10) until they stopped abruptly at random times. (E) Pointed ends of long filaments in the presence of Cdc12(FH1FH2)p grew continuously (solid line) at a rate of 0.6 ± 0.1 subunits/s (n = 10). Measurements from a single filament are indicated by filled circles. (F) Short filaments in the presence of Cdc12(FH1FH2)p. The total length of short filaments (arrowheads in C) versus total reaction time showed uniform growth at a rate of 0.3 ± 0.1 subunits/s (solid line; n = 10) and an apparent starting length of 240 ± 30 subunits. Diffraction increased the apparent 1/e width and length of individual actin filaments to 0.7 μm, or 260 subunits, so the starting length was zero.
Figure 3.
Real-time visualization of actin assembly in the presence of Cdc12(FH1FH2)p or mouse capping protein by TIRF microscopy. The conditions were as follows: 3 μM (25% Oregon green 488–labeled) Mg-ATP actin, 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 0.5% methylcellulose at 25°C. Flow cells were coated with NEM-myosin to capture the actin filaments. Direct comparisons of samples stained with rhodamine-phalloidin and adsorbed to coverslips coated with poly-
l
-lysine (Fig. 4) showed that the conditions of these TIRF experiments strongly favored the observation of long filaments. In fact, we observed no filaments at concentrations of Cdc12(FH1FH2)p >500 nM, where the biochemical results (and rhodamine-phalloidin staining) indicated numerous short filaments. (A–C) Images of actin filaments 500 s after initiating the reactions. Bar, 10 μm or 3,700 subunits. (A) Actin alone. (B) Actin with 50 nM _Mm_CP. Both long filaments (n = 15) and short filaments (n = 12; arrowheads) grew at a pointed end rate of 0.5 ± 0.1 subunits/s. (C) Actin with 250 nM Cdc12(FH1FH2)p. (D–F) Measurements of changes in filament length with time. Control filament barbed ends grew at 19.0 ± 0.1 subunits/s (n = 16 filaments), and pointed ends grew at 0.8 ± 0.1 subunits/s (n = 16). (D) Barbed ends in the presence of Cdc12(FH1FH2)p. The longer filaments (dashed line) grew at 25.2 ± 0.2 subunits/s (n = 10) until they stopped abruptly at random times. (E) Pointed ends of long filaments in the presence of Cdc12(FH1FH2)p grew continuously (solid line) at a rate of 0.6 ± 0.1 subunits/s (n = 10). Measurements from a single filament are indicated by filled circles. (F) Short filaments in the presence of Cdc12(FH1FH2)p. The total length of short filaments (arrowheads in C) versus total reaction time showed uniform growth at a rate of 0.3 ± 0.1 subunits/s (solid line; n = 10) and an apparent starting length of 240 ± 30 subunits. Diffraction increased the apparent 1/e width and length of individual actin filaments to 0.7 μm, or 260 subunits, so the starting length was zero.
Figure 4.
Effects of Cdc12(FH1FH2)p and mouse capping protein on spontaneous polymerization of Mg-ATP muscle actin. The buffer used was the same as in Fig. 2. (A and B) The time course of the polymerization of 4 μM Mg-ATP actin (5% pyrene labeled) was monitored by fluorescence in the presence of a range of concentrations of Cdc12(FH1FH2)p, (•) 0, (○) 5 nM, (▪) 25 nM, and (□) 500 nM, or a range of concentrations (nM) of _Mm_CP, (•) 0, (▴) 2.5 nM, (▵) 17.5 nM, and (♦) 250 nM. (B) Dependence of the concentration of apparent pointed ends ([Endsapp], nM) on the concentration of (•) Cdc12(FH1FH2)p or (○) _Mm_CP calculated from the rate of polymerization at the time where 40% (1.6 μM) of the actin was polymerized. (C) Effect of mixtures of Cdc12(FH1FH2)p and _Mm_CP on the time course of actin assembly; (•) 4 μM actin alone or 4 μM actin with (○) 10 nM Cdc12(FH1FH2)p, (▪) 4 nM _Mm_CP, (□) 10 nM Cdc12(FH1FH2)p + 4 nM _Mm_CP, (▴) 50 nM Cdc12(FH1FH2)p, (▵) 17 nM _Mm_CP, or (♦) 50 nM Cdc12(FH1FH2)p + 17 nM _Mm_CP. (D) Yield of [Endsapp] (calculated from B) per capping molecule as a function of the concentration of (•) Cdc12(FH1FH2)p or (○) _Mm_CP. (E–G) Fluorescence micrographs of filaments assembled to steady state from actin alone or with Cdc12(FH1FH2)p or _Mm_CP. Samples were labeled with rhodamine-phalloidin and adsorbed to glass coverslips coated with poly-
l
-lysine to capture short filaments (∼0.1–0.5 μm), but many more short filaments were free in solution. Bar, 10 μm. (E) 4 μM Mg-ATP actin assembled alone for 45 min. (F) Actin assembled with 100 nM _Mm_CP for 10 min. (G) Actin assembled with 200 nM Cdc12(FH1FH2)p for 10 min. (H) Time course of polymerization of the indicated concentrations of Mg-ATP actin alone (○) or with (□) 100 nM Cdc12(FH1FH2)p or (▵) 25 nM _Mm_CP.
Figure 4.
Effects of Cdc12(FH1FH2)p and mouse capping protein on spontaneous polymerization of Mg-ATP muscle actin. The buffer used was the same as in Fig. 2. (A and B) The time course of the polymerization of 4 μM Mg-ATP actin (5% pyrene labeled) was monitored by fluorescence in the presence of a range of concentrations of Cdc12(FH1FH2)p, (•) 0, (○) 5 nM, (▪) 25 nM, and (□) 500 nM, or a range of concentrations (nM) of _Mm_CP, (•) 0, (▴) 2.5 nM, (▵) 17.5 nM, and (♦) 250 nM. (B) Dependence of the concentration of apparent pointed ends ([Endsapp], nM) on the concentration of (•) Cdc12(FH1FH2)p or (○) _Mm_CP calculated from the rate of polymerization at the time where 40% (1.6 μM) of the actin was polymerized. (C) Effect of mixtures of Cdc12(FH1FH2)p and _Mm_CP on the time course of actin assembly; (•) 4 μM actin alone or 4 μM actin with (○) 10 nM Cdc12(FH1FH2)p, (▪) 4 nM _Mm_CP, (□) 10 nM Cdc12(FH1FH2)p + 4 nM _Mm_CP, (▴) 50 nM Cdc12(FH1FH2)p, (▵) 17 nM _Mm_CP, or (♦) 50 nM Cdc12(FH1FH2)p + 17 nM _Mm_CP. (D) Yield of [Endsapp] (calculated from B) per capping molecule as a function of the concentration of (•) Cdc12(FH1FH2)p or (○) _Mm_CP. (E–G) Fluorescence micrographs of filaments assembled to steady state from actin alone or with Cdc12(FH1FH2)p or _Mm_CP. Samples were labeled with rhodamine-phalloidin and adsorbed to glass coverslips coated with poly-
l
-lysine to capture short filaments (∼0.1–0.5 μm), but many more short filaments were free in solution. Bar, 10 μm. (E) 4 μM Mg-ATP actin assembled alone for 45 min. (F) Actin assembled with 100 nM _Mm_CP for 10 min. (G) Actin assembled with 200 nM Cdc12(FH1FH2)p for 10 min. (H) Time course of polymerization of the indicated concentrations of Mg-ATP actin alone (○) or with (□) 100 nM Cdc12(FH1FH2)p or (▵) 25 nM _Mm_CP.
Figure 4.
Effects of Cdc12(FH1FH2)p and mouse capping protein on spontaneous polymerization of Mg-ATP muscle actin. The buffer used was the same as in Fig. 2. (A and B) The time course of the polymerization of 4 μM Mg-ATP actin (5% pyrene labeled) was monitored by fluorescence in the presence of a range of concentrations of Cdc12(FH1FH2)p, (•) 0, (○) 5 nM, (▪) 25 nM, and (□) 500 nM, or a range of concentrations (nM) of _Mm_CP, (•) 0, (▴) 2.5 nM, (▵) 17.5 nM, and (♦) 250 nM. (B) Dependence of the concentration of apparent pointed ends ([Endsapp], nM) on the concentration of (•) Cdc12(FH1FH2)p or (○) _Mm_CP calculated from the rate of polymerization at the time where 40% (1.6 μM) of the actin was polymerized. (C) Effect of mixtures of Cdc12(FH1FH2)p and _Mm_CP on the time course of actin assembly; (•) 4 μM actin alone or 4 μM actin with (○) 10 nM Cdc12(FH1FH2)p, (▪) 4 nM _Mm_CP, (□) 10 nM Cdc12(FH1FH2)p + 4 nM _Mm_CP, (▴) 50 nM Cdc12(FH1FH2)p, (▵) 17 nM _Mm_CP, or (♦) 50 nM Cdc12(FH1FH2)p + 17 nM _Mm_CP. (D) Yield of [Endsapp] (calculated from B) per capping molecule as a function of the concentration of (•) Cdc12(FH1FH2)p or (○) _Mm_CP. (E–G) Fluorescence micrographs of filaments assembled to steady state from actin alone or with Cdc12(FH1FH2)p or _Mm_CP. Samples were labeled with rhodamine-phalloidin and adsorbed to glass coverslips coated with poly-
l
-lysine to capture short filaments (∼0.1–0.5 μm), but many more short filaments were free in solution. Bar, 10 μm. (E) 4 μM Mg-ATP actin assembled alone for 45 min. (F) Actin assembled with 100 nM _Mm_CP for 10 min. (G) Actin assembled with 200 nM Cdc12(FH1FH2)p for 10 min. (H) Time course of polymerization of the indicated concentrations of Mg-ATP actin alone (○) or with (□) 100 nM Cdc12(FH1FH2)p or (▵) 25 nM _Mm_CP.
Figure 5.
Visualization of actin filament elongation in the presence of profilin and Cdc12(FH1FH2)p. The buffer used was the same as in Fig. 3. (A–C) Fixed samples from TIRF microscopy reactions were labeled with rhodamine-phalloidin 15 min after the reaction start, diluted 1:59, and adsorbed to glass coverslips coated with poly-
l
-lysine. (A) 3 μM actin alone. (B) Actin plus 5 μM profilin. (C) Actin plus 100 nM Cdc12(FH1FH2)p and 5 μM profilin. Bar, 10 μm or 3,700 subunits. (D–I) Growth of individual actin filament ends was observed with TIRF microscopy. For actin alone, (D) pointed ends grew at 0.7 ± 0.1 subunits/s (n = 10) and (E) barbed ends grew at 16.2 ± 0.1 subunits/s (n = 10). In the presence of 5 μM profilin, (F) pointed end growth was inhibited to 0.0 ± 0.1 subunits/s (n = 11), whereas (G) barbed ends grew at 16.6 ± 0.1 subunits/s (n = 11). In the presence of 5 μM profilin and 100 nM Cdc12(FH1FH2)p, (H) pointed end growth was inhibited to 0.00 ± 0.03 subunits/s (n = 13), (I) whereas barbed ends grew at an initial rate of 12.4 ± 5.9 subunits/s (n = 12) before the free actin subunit pool was depleted. (I) The total length over time for each filament was fitted to an exponential growth curve to estimate the initial growth rate and nucleation time. Five example filaments are shown.
Figure 6.
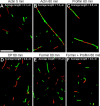
Effects of Cdc12(FH1FH2)p and profilin on actin filament annealing. Merged micrographs of red and green fluorescence. The buffer used was the same as in Fig. 2. Equal concentrations (0.25 μM) of red (rhodamine-phalloidin)- and green (Alexa® green–phalloidin)-labeled actin filaments were mixed, sheared through a 26-gauge needle, and allowed to anneal for up to 60 min before dilution and absorption to a coverslip coated with poly-
l
-lysine. Bar, 10 μm. (A) Actin filaments alone allowed to anneal for 5 min. (B) Actin filaments alone allowed to anneal for 60 min. (C) Actin filaments with 5 μM profilin after 60 min. (D) Actin filaments with 400 nM CP after 60 min. (E) Actin filaments with 400 nM Cdc12(FH1FH2)p after 60 min. (F) Actin filaments with 400 nM Cdc12(FH1FH2)p and 5 μM profilin after 60 min.
Figure 7.
Profilin inhibits capping of actin filament barbed ends by Cdc12(FH1FH2)p. The buffer used was the same as in Fig. 2. (A and B) Dependence of the polymerization of 1 μM actin on the concentration of Cdc12(FH1FH2)p and on the presence of profilin. 5 μM polymerized actin (10% pyrene labeled) was diluted to 1 μM in the presence of either (A) a range of Cdc12(FH1FH2)p concentrations and 5 μM profilin or (B) a range of profilin concentrations and 50 nM Cdc12(FH1FH2)p. After 16 h, the actin polymer concentration was measured by pyrene fluorescence. (A) Dependence of actin polymer concentration on the concentration of Cdc12(FH1FH2)p either (•) alone or Cdc12(FH1FH2)p with (▪) 5 μM wild-type profilin, (⋄) 5 μM profilin-Y5D, or (▵) 5 μM profilin-K81E. (B) Dependence of actin polymer concentration on the concentration of profilin in either the (•) absence or (▪) presence of 50 nM Cdc12(FH1FH2)p. (C and D) Time course of the depolymerization of 5 μM actin filaments (70% pyrene labeled) after dilution to 0.1 μM. (C) Depolymerization of (○) actin alone or actin in the presence of (•) 5 μM wild-type profilin, (⋄) 1 nM _Mm_CP, (♦) 1 nM _Mm_CP + 5 μM wild-type profilin, (▵) 5 nM _Mm_CP, or (▴) 5 nM _Mm_CP + 5 μM wild-type profilin. (D) Depolymerization of (○) actin alone or actin in the presence of (▴) 2 nM Cdc12(FH1FH2)p, (▾) 2 nM Cdc12(FH1FH2)p + 5 μM wild-type profilin, (▵) 2 nM Cdc12(FH1FH2)p + 5 μM profilin-Y5D, (▿) 2 nM Cdc12(FH1FH2)p + 5 μM profilin-K81E, (•) 10 nM Cdc12(FH1FH2)p, (▪) 10 nM Cdc12(FH1FH2)p + 5 μM wild-type profilin, (⋄) 10 nM Cdc12(FH1FH2)p + 5 μM profilin-Y5D, or (□) 10 nM Cdc12(FH1FH2)p + 5 μM profilin-K81E.
Figure 8.
Effect of profilin on spontaneous polymerization of Mg-ATP muscle actin in the presence of Cdc12(FH1FH2)p or mouse capping protein. The buffer used was the same as in Fig. 2. (A) Time course of the polymerization of 4 μM Mg-ATP actin (2% pyrene labeled) in the presence of Cdc12(FH1FH2)p and either wild-type or mutant profilins; (□) 4 μM Mg-ATP actin alone or with (○) 25 nM Cdc12(FH1FH2)p, (▿) 5 μM wild-type profilin, (•) 5 μM profilin-K81E with a reduced affinity for actin, (▵) 5 μM profilin-Y5D with a reduced affinity for poly-
l
-proline, (▴) 25 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin, (⋄) 25 nM Cdc12(FH1FH2)p and 5 μM profilin-K81E, or (♦) 25 nM Cdc12(FH1FH2)p and 5 μM profilin-Y5D. (B) Dependence of the concentration of ends produced by 4 μM Mg-ATP actin and 25 nM Cdc12(FH1FH2), as a function of the concentration of either (•) wild-type profilin or (○) profilin-Y5D. [Endsapp] was calculated at the time when 40% (1.6 μM) of the actin was polymerized, assuming barbed end growth. (C) Effect of profilin on capping protein nucleation. Time course of the polymerization of (•) 4 μM Mg-ATP actin alone or in the presence of (○) 5 μM wild-type profilin, (▪) 5 μM profilin-Y5D, (▵) 25 nM Cdc12(FH1FH2)p, (▴) 12.5 nM _Mm_CP, (♦) 25 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin, (□) 25 nM Cdc12(FH1FH2)p and 5 μM profilin-Y5D, (⋄) 12.5 nM _Mm_CP and 5 μM wild-type profilin, or (▿) 12.5 nM _Mm_CP and 5 μM profilin-Y5D. (D) Time course of the polymerization of the indicated concentrations of Mg-ATP actin alone (○) or with 25 nM Cdc12(FH1FH2)p alone (□), or with 25 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin (▵). Profilin lowered the concentration of actin monomer required for Cdc12(FH1FH2)p to stimulate polymerization. (E–H) Fluorescence micrographs of actin filaments labeled with rhodamine-phalloidin and adsorbed to glass coverslips coated with poly-
l
-lysine. Bar, 10 μm. (E) 4 μM Mg-ATP actin assembled alone. (F) Actin assembled with 5 μM wild-type profilin. (G) Actin assembled with 100 nM Cdc12(FH1FH2)p. (H) Actin assembled with 100 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin.
Figure 8.
Effect of profilin on spontaneous polymerization of Mg-ATP muscle actin in the presence of Cdc12(FH1FH2)p or mouse capping protein. The buffer used was the same as in Fig. 2. (A) Time course of the polymerization of 4 μM Mg-ATP actin (2% pyrene labeled) in the presence of Cdc12(FH1FH2)p and either wild-type or mutant profilins; (□) 4 μM Mg-ATP actin alone or with (○) 25 nM Cdc12(FH1FH2)p, (▿) 5 μM wild-type profilin, (•) 5 μM profilin-K81E with a reduced affinity for actin, (▵) 5 μM profilin-Y5D with a reduced affinity for poly-
l
-proline, (▴) 25 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin, (⋄) 25 nM Cdc12(FH1FH2)p and 5 μM profilin-K81E, or (♦) 25 nM Cdc12(FH1FH2)p and 5 μM profilin-Y5D. (B) Dependence of the concentration of ends produced by 4 μM Mg-ATP actin and 25 nM Cdc12(FH1FH2), as a function of the concentration of either (•) wild-type profilin or (○) profilin-Y5D. [Endsapp] was calculated at the time when 40% (1.6 μM) of the actin was polymerized, assuming barbed end growth. (C) Effect of profilin on capping protein nucleation. Time course of the polymerization of (•) 4 μM Mg-ATP actin alone or in the presence of (○) 5 μM wild-type profilin, (▪) 5 μM profilin-Y5D, (▵) 25 nM Cdc12(FH1FH2)p, (▴) 12.5 nM _Mm_CP, (♦) 25 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin, (□) 25 nM Cdc12(FH1FH2)p and 5 μM profilin-Y5D, (⋄) 12.5 nM _Mm_CP and 5 μM wild-type profilin, or (▿) 12.5 nM _Mm_CP and 5 μM profilin-Y5D. (D) Time course of the polymerization of the indicated concentrations of Mg-ATP actin alone (○) or with 25 nM Cdc12(FH1FH2)p alone (□), or with 25 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin (▵). Profilin lowered the concentration of actin monomer required for Cdc12(FH1FH2)p to stimulate polymerization. (E–H) Fluorescence micrographs of actin filaments labeled with rhodamine-phalloidin and adsorbed to glass coverslips coated with poly-
l
-lysine. Bar, 10 μm. (E) 4 μM Mg-ATP actin assembled alone. (F) Actin assembled with 5 μM wild-type profilin. (G) Actin assembled with 100 nM Cdc12(FH1FH2)p. (H) Actin assembled with 100 nM Cdc12(FH1FH2)p and 5 μM wild-type profilin.
Similar articles
- Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces.
Kovar DR, Pollard TD. Kovar DR, et al. Proc Natl Acad Sci U S A. 2004 Oct 12;101(41):14725-30. doi: 10.1073/pnas.0405902101. Epub 2004 Sep 17. Proc Natl Acad Sci U S A. 2004. PMID: 15377785 Free PMC article. - Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast.
Kovar DR, Wu JQ, Pollard TD. Kovar DR, et al. Mol Biol Cell. 2005 May;16(5):2313-24. doi: 10.1091/mbc.e04-09-0781. Epub 2005 Mar 2. Mol Biol Cell. 2005. PMID: 15743909 Free PMC article. - The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation.
Paul AS, Pollard TD. Paul AS, et al. Curr Biol. 2008 Jan 8;18(1):9-19. doi: 10.1016/j.cub.2007.11.062. Epub 2007 Dec 20. Curr Biol. 2008. PMID: 18160294 Free PMC article. - Formins: signaling effectors for assembly and polarization of actin filaments.
Evangelista M, Zigmond S, Boone C. Evangelista M, et al. J Cell Sci. 2003 Jul 1;116(Pt 13):2603-11. doi: 10.1242/jcs.00611. J Cell Sci. 2003. PMID: 12775772 Review. - Review of the mechanism of processive actin filament elongation by formins.
Paul AS, Pollard TD. Paul AS, et al. Cell Motil Cytoskeleton. 2009 Aug;66(8):606-17. doi: 10.1002/cm.20379. Cell Motil Cytoskeleton. 2009. PMID: 19459187 Free PMC article. Review.
Cited by
- Z-line formins promote contractile lattice growth and maintenance in striated muscles of C. elegans.
Mi-Mi L, Votra S, Kemphues K, Bretscher A, Pruyne D. Mi-Mi L, et al. J Cell Biol. 2012 Jul 9;198(1):87-102. doi: 10.1083/jcb.201202053. Epub 2012 Jul 2. J Cell Biol. 2012. PMID: 22753896 Free PMC article. - The role of the actin cytoskeleton in plant cell signaling.
Drøbak BK, Franklin-Tong VE, Staiger CJ. Drøbak BK, et al. New Phytol. 2004 Jul;163(1):13-30. doi: 10.1111/j.1469-8137.2004.01076.x. New Phytol. 2004. PMID: 33873778 Review. - The F-actin bundler α-actinin Ain1 is tailored for ring assembly and constriction during cytokinesis in fission yeast.
Li Y, Christensen JR, Homa KE, Hocky GM, Fok A, Sees JA, Voth GA, Kovar DR. Li Y, et al. Mol Biol Cell. 2016 Jun 1;27(11):1821-33. doi: 10.1091/mbc.E16-01-0010. Epub 2016 Apr 13. Mol Biol Cell. 2016. PMID: 27075176 Free PMC article. - F-Actin Cytoskeleton Network Self-Organization Through Competition and Cooperation.
Kadzik RS, Homa KE, Kovar DR. Kadzik RS, et al. Annu Rev Cell Dev Biol. 2020 Oct 6;36:35-60. doi: 10.1146/annurev-cellbio-032320-094706. Annu Rev Cell Dev Biol. 2020. PMID: 33021819 Free PMC article. Review. - Cdk1 phosphorylation of fission yeast paxillin inhibits its cytokinetic ring localization.
Mangione MC, Chen JS, Gould KL. Mangione MC, et al. Mol Biol Cell. 2021 Aug 15;32(17):1534-1544. doi: 10.1091/mbc.E20-12-0807. Epub 2021 Jun 16. Mol Biol Cell. 2021. PMID: 34133210 Free PMC article.
References
- Alberts, A.S. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276:2824–2830. - PubMed
- Andrianantoandro, E., L. Blanchoin, D. Sept, J.A. McCammon, and T.D. Pollard. 2001. Kinetic mechanism of end-to-end annealing of actin filaments. J. Mol. Biol. 312:721–730. - PubMed
- Balasubramanian, M.K., D. McCollum, and K.L. Gould. 1997. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 283:494–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM-26338/GM/NIGMS NIH HHS/United States
- R37 GM026132/GM/NIGMS NIH HHS/United States
- GM-26132/GM/NIGMS NIH HHS/United States
- R01 GM026338/GM/NIGMS NIH HHS/United States
- R01 GM026132/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous