Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex - PubMed (original) (raw)
Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex
Manuela E Hase et al. Mol Biol Cell. 2003 May.
Erratum in
- Mol Biol Cell. 2003 Aug;14(8):preceding Table of Contents
Abstract
Tpr is a 267-kDa protein forming coiled coil-dominated homodimers that locate at the nucleoplasmic side of the nuclear pore complex (NPC). The proteins that tether Tpr to this location are unknown. Moreover, the question whether Tpr itself might act as a scaffold onto which other NPC components need to be assembled has not been answered to date. To assess Tpr's role as an architectural element of the NPC, we have studied the sequential disassembly and reassembly of NPCs in mitotic cells, paralleled by studies of cells depleted of Tpr as a result of posttranscriptional tpr gene silencing by RNA interference (RNAi). NPC assembly and recruitment of several nucleoporins, including Nup50, Nup93, Nup96, Nup98, Nup107, and Nup153, in anaphase/early telophase is shown to precede NPC association of Tpr in late telophase. In accordance, cellular depletion of Tpr by RNAi does not forestall binding of these nucleoporins to the NPC. In a search for proteins that moor Tpr to the NPC, we have combined the RNAi approach with affinity-chromatography and yeast two-hybrid interaction studies, leading to the identification of nucleoporin Nup153 as the binding partner for Tpr. The specificity of this interaction is demonstrated by its sensitivity to Tpr amino acid substitution mutations that abolish Tpr's ability to adhere to the NPC and affect the direct binding of Tpr to Nup153. Accordingly, cellular depletion of Nup153 by RNAi is shown to result in mislocalization of Tpr to the nuclear interior. Nup153 deficiency also causes mislocalization of Nup50 but has no direct effect on NPC localization of the other nucleoporins studied in this investigation. In summary, these results render Tpr a protein only peripherally attached to the NPC that does not act as an essential scaffold for other nucleoporins.
Figures
Figure 1.
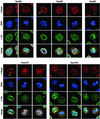
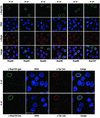
Orderly disassembly of presumptive basket nucleoporins and Tpr in prophase. HeLa cells were fixed with formaldehyde and double-labeled with mouse or rabbit antibodies against Tpr (green staining), together with rabbit or guinea-pig antibodies against aa 126–147 of hNup50, aa 1743–1763 of hNup196 corresponding to the C-terminus of the proteolytic Nup96 product, aa 596–618 of hNup98, aa 33–51 of hNup107, and aa 21–36 of hNup153 (red staining). DNA staining with TO-PRO-3 is shown in blue. Dissociation of Tpr from the nuclear periphery of prophase cells is preceded by early loss of Nup50 and Nup98, but occurs before late disassembly of Nup107. Examples of cells in mid and late prophase/early prometaphase are labeled with arrows. Arrowheads demark a striking redistribution of Nup96 and Nup107 to the proximity of the centrosomes in cells early in prophase. Bars, 10 μm; for all micrographs.
Figure 2.
Sequential recruitment of presumptive basket nucleoporins and Tpr into the reassembling NPC at the end of mitosis. HeLa cells were fixed with formaldehyde and double-labeled with antibodies against Tpr, Nup50, Nup96, Nup98, Nup107, and Nup153 as in Figure 1. Nup93 was labeled with antibodies against the recombinant N-terminus of hNup93 (B) and aa 350–369 of hNup93 (C). (A and B) Nucleoporins are recruited back to the periphery of the segregated chromatids in anaphase (A), whereas Tpr remains homogeneously dispersed throughout the cytoplasm until late in telophase (B). (C) Nucleoporins such as Nup93 are stably linked to the nuclear periphery in telophase cells and withstand detergent extraction of cells before fixation. Tpr, in contrast, is near-quantitatively extracted from such cells (arrows in left panel). Tpr is reincorporated into nuclei only late in telophase/early in G1, paralleling the decondensation of chromatin (arrowheads in right panel). Bar, 10 μm; for all micrographs.
Figure 3.
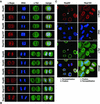
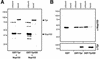
Posttranscriptional tpr gene silencing leads to cellular depletion of protein Tpr which neither alters NPC distribution nor prevents synthesis and NPC-association of other presumptive basket nucleoporins. (A) Immunoblotting of total cell proteins from control (–) and siRNA-treated (+) HeLa cells harvested 1, 3, and 4 d (d) posttransfection. Proteins from corresponding pairs of control and RNAi cells were loaded in near-equal amounts. Filters with identical loadings prepared in parallel were immunolabeled with rabbit antibodies against the recombinant C-terminus of Tpr, mAb PF190x7A8 against Nup153, and guinea-pig antibodies against aa 1784–1803 of hNup205, aa 33–51 of hNup107, 206–219 of hNup98, aa 993-1009 of hNup196 (Nup96), and aa 371–389 of hNup93. Note that the total amount of Tpr in a siRNA-treated cell population reaches a minimum after three to four days; the residual amount of Tpr mainly reflects a subpopulation of nontransfected cells (Kuznetsov et al., 2002). In contrast, no reduction is seen for the other nucleoporins. (B) IF microscopy of HeLa cells 3 or 4 d after transfection with Tpr siRNA duplexes. Cells were fixed with formaldehyde and double-labeled with antibodies against Tpr and the other nucleoporins as in Figure 1. Cells labeled with antibodies against Nup93 were permeabilized with detergent before fixation. Note that in populations transfected with siRNA duplexes, most cells show only traces of Tpr staining at the nuclear periphery; the usually bright nuclear rim staining is visible only in a minor subpopulation of cells that have remained untransfected. Tpr deficiency, however, neither inhibits the recruitment of other nucleoporins to the nuclear periphery nor affects the normal distribution of NPCs within the NE. Bar, 20 μm.
Figure 4.
Chemical cross-linking reveals physical proximity between presumptive basket nucleoporins and Tpr. HeLa cells were treated with or without thiol-cleavable cross-linker DTSSP and then solubilized with ionic detergents. The resulting extracts were immunoprecipitated with Tpr antibodies, and bound vs. unbound proteins analyzed by immunoblotting. Gels were loaded with the same percentage of each fraction. The left lanes show total cell extracts as reference. Relative masses of marker proteins in kDa are given at the left margin. The disulfide bond of DTSSP was reduced by treatment with β-mercaptoethanol before SDS-PAGE. Note that nucleoporins located at the nuclear side of the NPC coprecipitate with Tpr, whereas nucleoporin p62 located within the central pore channel is not.
Figure 5.
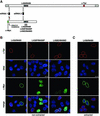
NPC binding of recombinant Tpr in cells silenced for wild-type tpr expression can be abolished by aa substitution mutations. (A) Silent point mutations introduced into the nt sequence encompassing the start codon of Tpr cDNAs protect from being targeted by siRNA duplexes that mediate wild-type mRNA degradation: Schematic presentation of the wild-type Tpr mRNA and one recombinant Tpr mRNA variant, here encoding aa 1–774 fused to an N-terminal Myc-tag (green box) and a C-terminal NLS. Relative positions of siRNA duplexes, both complementary to the wild-type sequence, are represented as boxes 1 and 2. Duplex 2 triggers degradation of both the wild-type and the recombinant mRNA. In contrast, duplex 1 affects only the wild-type mRNA since silent nt exchanges that prevent the RNAi initiation step were introduced into the recombinant sequence. In addition, 5′-UTR sequences (light gray box) identical to the wild-type mRNA and upstream of the silent mutations have been removed. Arrowheads demark relative positions of substitution mutations at L458 and M489 located within the NPC binding domain of Tpr (NBD). The red box stands for the Tpr sequence encoding the epitope recognized by α-Tpr aa 2063–2084. (B) Expression of Myc-tagged Tpr 1–774 and corresponding mutants with double proline or aspartic acid substitutions at aa 458 and aa 489 in HeLa cells transfected with Tpr siRNA duplex no.1 and consequently depleted of endogenous wild-type Tpr. Normal levels of wild-type Tpr in nontransfected cells shown as reference were detected with rabbit antibodies against Tpr aa 2063–2084 (red); recombinant Tpr was stained with mAb 9E10 against the N-terminal Myc-tag (green), and DNA with TO-PRO-3. Note that in wild-type Tpr-deficient cells only recombinant Tpr polypeptides with an intact NBD associate with the NE. In contrast, double substitution mutations at aa 458 and aa 489 abolish NPC binding and cause intranuclear accumulation. Confocal micrographs are of cells fixed with FA before permeabilization with detergents. (C) Expression of Myc-tagged Tpr 1–774 in wild-type Tpr-deficient cells. Before FA-fixation and labeling with antibodies, cells were extracted with Triton X-100. Bars, 10 μm.
Figure 6.
Sequence integrity of Tpr's NPC binding domain mediates specific binding to Nup153. GST fusion proteins encompassing the NBD of wild-type Tpr (GST-Tpr 172–651), the corresponding mutant with aspartic acid substitutions at aa 458 and aa 489 (GST-Tpr 172–651 DD) and GST alone were immobilized on glutathione Sepharose beads. After incubation with Hela cell extracts, bound and unbound proteins were analyzed by immunoblotting. Unbound material representing the initial column run-through was loaded in lane 1, material from column washes with PBS in lane 2, and material eluted with 1 M MgCl2 in PBS in lane 3. Final elutions with 10 mM and 20 mM reduced glutathione are shown in lane 4 and 5. (A) Immunoblots with mAb PF190x7A8 against Nup 153. The second immunoreactive polypeptide of lower molecular mass is considered being a product of Nup153 degradation during extract incubation. (B) Immunoblots with antibodies against aa 1–21 and aa 126–147 of hNup50, aa 371–389 of hNup93, aa 1743–1763 of hNup196 (Nup96), aa 206–219 of hNup98, aa 33–51 of hNup107, and aa 1784–1803 of hNup205. Here only the results with GST-Tpr 172–651 are shown. Note that salt-resistant protein-protein interactions only occur between Nup153 and the intact NBD of Tpr, but not between the NBD and other nucleoporins.
Figure 7.
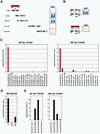
Yeast two-hybrid analysis reveals specific interaction between Tpr's NPC binding domain and a segment of the N-terminal domain of Nup153 in vivo. (A and B) Schemes of Tpr and Nup153 fragments, and representative results of β-galactosidase filter lift assays. Here, interactions between Nup153 polypeptide 228–611 fused to yeast GAL4-BD and Tpr segments fused to GAL4-AD are shown. (A) LacZ reporter gene activation only occurs upon coexpression of Nup153 and those Tpr segments that contain the region known to be essential for NPC binding (∼ aa 450–550; Hase et al., 2001). Filters shown were prepared in parallel, with settings for color reproduction being identical. (B) Tpr aa substitution mutations that do not impair homodimerization but inhibit binding to the NPC, abolish the interaction with Nup153. (C) Quantification of two-hybrid interactions between Tpr and nucleoporins by liquid β-galactosidase assays. Sequences of Nup 50, Nup93, Nup 96, Nup 98, Nup107, Nup153, and Nup205 were fused to GAL4-BD, and Tpr aa 172–651 to GAL4-AD, and vice versa. The vertical scale represents units of β-galactosidase activity. Each value represents the average of enzyme activity measured for three independent yeast clones; a minor background of β-galactosidase activity measured for cells coexpressing GAL4-AD and GAL4-BD alone was subtracted from each data set. (D) Quantification of two-hybrid interactions between Nup153 and Tpr's intact NBD, Tpr 172–651 DD, or Tpr 172–651 PP. (E) Quantification of two-hybrid interactions between Tpr's intact NBD and different segments of Nup153.
Figure 8.
Direct interaction of Nup153 and Tpr in vitro. (A) GST-Tpr 172-651 and GST-Tpr 172-651 DD were first incubated together with recombinant Nup 153 polypeptide 228–611 and then immobilized on glutathione Sepharose beads. Bound and unbound proteins were separated by SDS-PAGE and stained by Coomassie Blue. Arrows mark bands of Nup153 and Tpr, respectively. Asterisks mark one of several contaminating bacterial proteins not removed in the course of Nup153 purification. (B) Immunoblot identification of bound and unbound Nup153 proteins after incubation with same molar amounts of GST alone, GST-Tpr 172-651, and GST-Tpr 172-651 DD. The recombinant Tpr polypeptide was immunodetected with rabbit antibodies against Tpr aa 604–615. Lanes with bound proteins were loaded at eight times higher ratio of fraction than lanes with unbound material.
Figure 9.
Posttranscriptional Nup153 gene silencing leads to cellular depletion of protein Nup153 and nuclear redistribution of Tpr. IF microscopy of HeLa cells four days after transfection with Nup153 siRNA duplexes. Cells were either fixed with formaldehyde first, and then permeabilized with detergent (F→P), or permeabilized before fixation (P→F). Double-labeling was with mouse, rabbit, or guinea-pig antibodies as in Figure 1. In populations transfected with siRNA duplexes, most cells show only traces or no Nup153 staining at the nuclear periphery; the usually bright nuclear rim staining is visible only in a minor subpopulation of cells that have remained untransfected. Nup153 deficiency neither inhibits the recruitment of Nup93, Nup96, Nup98, and Nup107 to the nuclear periphery nor affects the normal distribution of NPCs within the NE, but strikingly impairs NPC-binding of Nup50 and Tpr. Normal NE-staining for Nup50 (arrows) is only seen in Nup153-positive cells; Nup50 released from the NE in Nup153-deficient cells often appears to add to the large pool of Nup50 generally found distributed throughout the nuclear interior. Such intranuclear forms of Nup50 can be near-quantitatively extracted by brief detergent treatment. Tpr molecules that have been prevented from binding to the NE in Nup153-deficient cells tend to form intranuclear aggregates that are detectable with rabbit antibodies (rb) against the C-terminal tail as well as with mAb (m) 203-37 against the rod domain of Tpr. Nup153 was labeled with either polyclonal guinea-pig antibodies (gp) against the N-terminal region, or with mAb PF190x7A8 against an epitope located between aa 439 and 611. Bars, 20 μm.
Similar articles
- Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket.
Krull S, Thyberg J, Björkroth B, Rackwitz HR, Cordes VC. Krull S, et al. Mol Biol Cell. 2004 Sep;15(9):4261-77. doi: 10.1091/mbc.e04-03-0165. Epub 2004 Jun 30. Mol Biol Cell. 2004. PMID: 15229283 Free PMC article. - Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA.
Rajanala K, Nandicoori VK. Rajanala K, et al. PLoS One. 2012;7(1):e29921. doi: 10.1371/journal.pone.0029921. Epub 2012 Jan 13. PLoS One. 2012. PMID: 22253824 Free PMC article. - Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization.
Hase ME, Kuznetsov NV, Cordes VC. Hase ME, et al. Mol Biol Cell. 2001 Aug;12(8):2433-52. doi: 10.1091/mbc.12.8.2433. Mol Biol Cell. 2001. PMID: 11514627 Free PMC article. - Function and assembly of nuclear pore complex proteins.
Bodoor K, Shaikh S, Enarson P, Chowdhury S, Salina D, Raharjo WH, Burke B. Bodoor K, et al. Biochem Cell Biol. 1999;77(4):321-9. Biochem Cell Biol. 1999. PMID: 10546895 Review. - Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.
Hamed M, Antonin W. Hamed M, et al. Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review.
Cited by
- Multiple conserved domains of the nucleoporin Nup124p and its orthologs Nup1p and Nup153 are critical for nuclear import and activity of the fission yeast Tf1 retrotransposon.
Sistla S, Pang JV, Wang CX, Balasundaram D. Sistla S, et al. Mol Biol Cell. 2007 Sep;18(9):3692-708. doi: 10.1091/mbc.e06-12-1062. Epub 2007 Jul 5. Mol Biol Cell. 2007. PMID: 17615301 Free PMC article. - Nucleocytoplasmic transport: integrating mRNA production and turnover with export through the nuclear pore.
Dimaano C, Ullman KS. Dimaano C, et al. Mol Cell Biol. 2004 Apr;24(8):3069-76. doi: 10.1128/MCB.24.8.3069-3076.2004. Mol Cell Biol. 2004. PMID: 15060131 Free PMC article. Review. No abstract available. - Spelling out the roles of individual nucleoporins in nuclear export of mRNA.
Tingey M, Li Y, Yu W, Young A, Yang W. Tingey M, et al. Nucleus. 2022 Dec;13(1):170-193. doi: 10.1080/19491034.2022.2076965. Nucleus. 2022. PMID: 35593254 Free PMC article. Review. - Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor.
Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. Rodriguez-Bravo V, et al. Cell. 2014 Feb 27;156(5):1017-31. doi: 10.1016/j.cell.2014.01.010. Cell. 2014. PMID: 24581499 Free PMC article. - Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export.
Fasken MB, Stewart M, Corbett AH. Fasken MB, et al. J Biol Chem. 2008 Oct 3;283(40):27130-43. doi: 10.1074/jbc.M803649200. Epub 2008 Aug 5. J Biol Chem. 2008. PMID: 18682389 Free PMC article.
References
- Arlucea, J., Andrade, R., Alonso, R., and Arechaga, J. (1998). The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J. Struct. Biol. 124, 51–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous