NSD1 is essential for early post-implantation development and has a catalytically active SET domain - PubMed (original) (raw)
NSD1 is essential for early post-implantation development and has a catalytically active SET domain
Geetha Vani Rayasam et al. EMBO J. 2003.
Abstract
The nuclear receptor-binding SET domain-containing protein (NSD1) belongs to an emerging family of proteins, which have all been implicated in human malignancy. To gain insight into the biological functions of NSD1, we have generated NSD1-deficient mice by gene disruption. Homozygous mutant NSD1 embryos, which initiate mesoderm formation, display a high incidence of apoptosis and fail to complete gastrulation, indicating that NSD1 is a developmental regulatory protein that exerts function(s) essential for early post-implantation development. We have also examined the enzymatic potential of NSD1 and found that its SET domain possesses intrinsic histone methyltransferase activity with specificity for Lys36 of histone H3 (H3-K36) and Lys20 of histone H4 (H4-K20).
Figures
Fig. 1. Targeted disruption of NSD1 using the Cre-loxP strategy. (A) Diagram showing a partial map of the genomic locus surrounding the NSD1 exon 2 (E2), the targeting construct, and the targeted allele before (L3) and after (L2 and _L_–) Cre-mediated excision of the neomycin resistance selection marker (Neo). The 5′ and 3′ probes used for Southern blot analyses and the fragment sizes detected with the 5′ probe upon _Xba_I digestion and with the 3′ probe upon _Hin_dIII digestion are indicated. Relevant restriction sites: X, _Xba_I; K, _Kpn_I; E, _Eco_RI; H, _Hin_dIII; S, _Sma_I; A, _Afl_II; Sp, _Spe_I. (B) Schematic representation of wild-type (WT) and mutant NSD1 proteins. The structural and functional domains are indicated. The putative product of the deleted NSD1 gene corresponds to a C-terminally truncated protein consisting of the first 310 amino acids of NSD1. (C) Southern blot analysis of DNAs derived from wild-type (P1) and targeted (BT157 and BT259) ES cells. Genomic DNA was digested with _Hin_dIII, blotted and hybridized with the 3′ probe. (D) Southern blot analysis of ES cell subclones using the 5′ probe, after Cre-mediated excision in BT259 _NSD1_L3/+ ES cells.
Fig. 2. Morphology of wild-type and _NSD1_–/– mutant embryos at E7.5 and E8.0. (A) PCR strategy for amplification of wild-type (WT) and excised (L–) NSD1 alleles. DNA samples were subjected to PCR amplification using a mixture of three primers (see Materials and methods). PCR amplification of the wild-type NSD1 allele by sense and antisense primers ZB197 and AAW199 produces a 550 bp DNA fragment (B, upper band), while PCR amplification of the excised allele by sense and antisense primers ZB197 and ZB200 produces a 350 bp DNA fragment (B, lower band). (B) Representative genotypic analysis of E7.5 embryos from an NSD1+/– intercross. (C) Dissected wild-type (left) and _NSD1_–/– mutant (right) littermates at E7.5 and E8.0. Abbreviations: ee, epiblast; ep, ectoplacental cone; h, head folds.
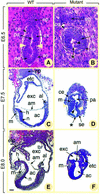
Fig. 3. Histological sections of normal (A, C and E) and presumptive _NSD1_–/– (B, D and F) embryos at E6.5 (A and B), E7.5 (C and D) and E8.0 (E and F). In (A) and (B), deciduas were kept intact. In (C–F), embryos were dissected out of the decidua. Yellow arrowheads in (A) and (D) show the boundary between embryonic and extraembryonic regions. Asterisks in (B) and (D) indicate gaps in the mutant epiblast, and black arrowheads point to pyknotic cells in the proamniotic cavity. Abbreviations: ac, amniotic cavity; al, allantois; am, amnion; ce, cuboidal visceral endoderm; e, ectoderm; ee, epiblast; ep, ectoplacental cone; etc, ectoplacental cavity; ex, extraembryonic ectoderm; exc, exocoelom; h, head folds; m, mesoderm; n, neurectoderm; pa, proamniotic cavity; se, squamous visceral endoderm. Scale bar: 20 µm (A and B); 70 µm (C–F).
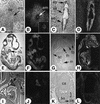
Fig. 4. TUNEL and in situ hybridization analyses of E7.5 normal and mutant embryos. Sections of E7.5 normal embryos (A, C, E, G, I and K) and their presumptive mutant littermates (B, D, F, H, J and L) were subjected to TUNEL reaction (A and B) and were hybridized with Brachyury (C and D), Shh (E and F), Twist (G and H), Hoxa1 (I and J), and Hoxb1 (K and L) antisense probes. In (A) and (B), brown-stained nuclei indicate end incorporation in DNA (arrowheads). Abbreviations: a, axial mesendoderm; al, allantois; h, head folds; m, mesoderm; no, node; s, primitive streak. Scale bar: 60 µm.
Fig. 5. In situ hybridization analysis of NSD1 transcript distribution at various stages of mouse development. Brightfield and darkfield views of the histological sections are shown side by side (left and right panel, respectively), revealing the signal grain as white dots. (A and B) E5.5 embryo sectioned in utero. Insert panels show magnification of the embryo. (C and D) Ubiquitous expression of NSD1 in the ectoplacental cone (ep), extraembryonic (ex) and embryonic (em) germ layers of an E7.5 embryo. (E and F) Ubiquitous NSD1 expression in an E9.5 embryo. ba, branchial arches; br, brain; sc, spinal cord. (G and H) Enhanced expression of NSD1 in the brain (br), intestine (in), spinal cord (sc), thymus (th) and tooth buds (tb) of an E16.5 fetus. (I and J) Detail of the E16.5 heart (ht), thymus (th) and salivary gland area (sg). (K and L) Section through the ossification center of the femur. bo, bone tissue; ca, cartilage (chondrocytes); pe, periosteum.
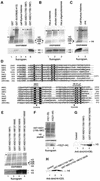
Fig. 6. The NSD1 SET domain has intrinsic HMTase activity with specificity for H3-K36 and H4-K20. (A) Coomassie blue-stained SDS–polyacrylamide gel and corresponding flurorogram of HMTase assays with the indicated GST fusion proteins and a mixture of purified calf thymus histones as substrates. (B) HMTase assays as in (A), except that reactions contained either a mixture of native core histones from HeLa cells (lane 1) or chicken (lane 2), or oligonucleosomes purified from HeLa cells (lane 3), and GST–NSD1(1700–1987) as enzyme. (C) HMTase assays as in (A), except that reactions contained either purified calf thymus histones (lane 1) or recombinant H4 (lane 2), and GST–NSD1(1700–1987) as enzyme. (D) Amino acid alignment of the NSD1 SET domain with other selected SET domains. The sequences were aligned using the CLUSTAL W program and manual adjustment. Invariant amino acids are highlighted in blue. Amino acids conserved in >60% of the proteins are highlighted in yellow. The two conserved amino acid stretches important for the methyltransferase catalytic activity of the SET domain are indicated. Mutations described in this study are indicated below the alignment. (E) The C1920S mutation generated a hyperactive SET domain mutant. Equal amounts of wild-type and mutant GST–NSD1 proteins (top panel) were compared for their HMTase activities (bottom panel) using equal amounts of purified calf thymus histones. (F) HMTase assays as in (A) using GST–NSD1(1700–1987) as enzyme and the indicated N-terminal peptides of H3. (G) Western blot analyses, using anti-dim(H3-K36), of HMTase assays containing recombinant H3 and the indicated GST–NSD1 proteins. (H) Western blot analyses, using anti-dim(H4-K20), of HMTase assays containing recombinant H4 and the indicated GST–NSD1 proteins.
Similar articles
- NSD1 PHD domains bind methylated H3K4 and H3K9 using interactions disrupted by point mutations in human sotos syndrome.
Pasillas MP, Shah M, Kamps MP. Pasillas MP, et al. Hum Mutat. 2011 Mar;32(3):292-8. doi: 10.1002/humu.21424. Hum Mutat. 2011. PMID: 21972110 - Nizp1, a novel multitype zinc finger protein that interacts with the NSD1 histone lysine methyltransferase through a unique C2HR motif.
Nielsen AL, Jørgensen P, Lerouge T, Cerviño M, Chambon P, Losson R. Nielsen AL, et al. Mol Cell Biol. 2004 Jun;24(12):5184-96. doi: 10.1128/MCB.24.12.5184-5196.2004. Mol Cell Biol. 2004. PMID: 15169884 Free PMC article. - NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis.
Wang GG, Cai L, Pasillas MP, Kamps MP. Wang GG, et al. Nat Cell Biol. 2007 Jul;9(7):804-12. doi: 10.1038/ncb1608. Epub 2007 Jun 24. Nat Cell Biol. 2007. PMID: 17589499 - Structure of SET domain proteins: a new twist on histone methylation.
Marmorstein R. Marmorstein R. Trends Biochem Sci. 2003 Feb;28(2):59-62. doi: 10.1016/S0968-0004(03)00007-0. Trends Biochem Sci. 2003. PMID: 12575990 Review. - Unsafe SETs: histone lysine methyltransferases and cancer.
Schneider R, Bannister AJ, Kouzarides T. Schneider R, et al. Trends Biochem Sci. 2002 Aug;27(8):396-402. doi: 10.1016/s0968-0004(02)02141-2. Trends Biochem Sci. 2002. PMID: 12151224 Review.
Cited by
- Sound of silence: the properties and functions of repressive Lys methyltransferases.
Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Mozzetta C, et al. Nat Rev Mol Cell Biol. 2015 Aug;16(8):499-513. doi: 10.1038/nrm4029. Nat Rev Mol Cell Biol. 2015. PMID: 26204160 Review. - Transcription-associated histone modifications and cryptic transcription.
Smolle M, Workman JL. Smolle M, et al. Biochim Biophys Acta. 2013 Jan;1829(1):84-97. doi: 10.1016/j.bbagrm.2012.08.008. Epub 2012 Sep 7. Biochim Biophys Acta. 2013. PMID: 22982198 Free PMC article. Review. - Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila.
Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, Garza D, Peters AH, Schübeler D. Bell O, et al. EMBO J. 2007 Dec 12;26(24):4974-84. doi: 10.1038/sj.emboj.7601926. Epub 2007 Nov 15. EMBO J. 2007. PMID: 18007591 Free PMC article. - A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin.
Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. Schotta G, et al. Genes Dev. 2004 Jun 1;18(11):1251-62. doi: 10.1101/gad.300704. Epub 2004 May 14. Genes Dev. 2004. PMID: 15145825 Free PMC article. - Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana.
Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. Xu L, et al. Mol Cell Biol. 2008 Feb;28(4):1348-60. doi: 10.1128/MCB.01607-07. Epub 2007 Dec 10. Mol Cell Biol. 2008. PMID: 18070919 Free PMC article.
References
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. - PubMed
- Angrand P.O., Apiou,F., Stewart,A.F., Dutrillaux,B., Losson,R. and Chambon,P. (2001) NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics, 74, 79–88. - PubMed
- Beisel C., Imhof,A., Greene,J., Kremmer,E. and Sauer,F. (2002) Histone methylation by the Drosophila epigenetic regulator Ash1. Nature, 419, 857–862. - PubMed
- Chesi M., Nardini,E., Lim,R.S., Smith,K.D., Kuehl,W.M. and Bergsagel,P.L. (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood, 92, 3025–3034. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials