The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling - PubMed (original) (raw)
The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling
Heather Szerlong et al. EMBO J. 2003.
Abstract
Nuclear actin-related proteins (ARPs) are essential components of chromatin remodeling and modifying complexes, but their functions and relationship to actin remain elusive. The yeast SWI/SNF and RSC complexes contain Arp7 and Arp9, and are shown to form a stable heterodimer with the properties of a functional module. Arp7 and Arp9 rely on their actin-related regions for heterodimerization, and their unique C-termini cooperate for assembly into RSC. We suggest that regulated ARP-ARP (and possibly ARP-beta-actin) heterodimerization might be a conserved feature of chromatin complexes. A RSC complex lacking Arp7/9 was isolated that displays robust nucleosome remodeling activity, suggesting a separate essential role for ARPs in the regulation of chromatin structure. A screen for suppressors of arp mutations yielded the DNA bending architectural transcription factor Nhp6, which interacts with RSC complex physically and functionally and shows facilitated binding to nucleosomes by RSC. We propose that Arp7/9 dimers function with DNA bending proteins to facilitate proper chromatin architecture and complex- complex interactions.
Figures
Fig. 1. New conditional mutations in ARP7 and ARP9. (A) Mutations in arp7 (red) and arp9 (green) superimposed on the structure of actin (Kabsch et al., 1990). (B) C-terminal regions lacking in arp7 (red) or arp9 (green) truncation alleles. Regions of similarity are boxed in grey. Rabbit actin (top row). (C) Growth of Ts– arp7 and arp9 strains. Strains given below. (D) Protein expression profiles of arp proteins. Whole-cell extracts from WT and Ts– arp strains grown at the temperatures indicated were immunoblotted with either polyclonal anti-Arp7 or anti-Arp9 antibody. Strains: pNCT.ARP7 (YBC1533), pNCT arp7E411K (YBC776), pNCT.arp7S33F (YBC788), pNCT.arp7_Δ_C1 (YBC786), pNCT.arp7_Δ_C2 (YBC1534); for ARP9, pNCT.ARP9 (YBC1535), pNCT.arp9-1 (YBC1536) or pNCT.arp9_Δ_C (YBC775). Wild type (WT) (YBC605).
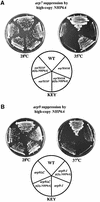
Fig. 2. ARP7 and ARP9 cosuppression relationships and suppression by mra1. (A) Suppression of arp7 mutations by multicopy ARP9. Strains: WT pNCT.ARP7 (YBC1533). arp7 Ts– alleles: pNCT.arp7S33F (YBC788), pNCT arp7E411K (YBC776), pNCT.arp7_Δ_C2(YBC1534). Growth of WT and arp7 Ts– strains transformed with either YEp24.ARP9 or YEp24 alone on SC-Ura at 28°C or 36°C. (B) Suppression of arp9 mutations by multicopy ARP7. Growth of WT and arp9 Ts– strains transformed with either YEp24.ARP7 or YEp24 alone on SC-Ura at 28°C or 37°C. Strains: WT (YBC1535), pNCT.arp9-1 (YBC1536) and pNCT.arp9_Δ_C (YBC775). (C) mra1 is a suppressor of _arp7_Δ and _arp9_Δ mutations. All W303 background: WT (BCY405), _arp9_Δ _arp7_Δ (BCY393) and the triple _arp9_Δ _arp7_Δ mra1-1 (BCY395) were compared for growth on rich media plates at 30°C or 38°C.
Fig. 3. Purification and characterization of a stable Arp7–Arp9 heterodimer. (A) Arp7 and Arp9 copurify. Histidine-tagged Arp7 and Flag-tagged Arp9 were coexpressed in E.coli using a bi-cistronic vector (lane 1). Soluble protein extracts were subjected to tandem affinity purification using Ni-NTA resin (lane 2) followed by anti-Flag resin (lane 3). Samples were separated in a 7.5% acrylamide–SDS gel and visualized by staining with Coomassie Blue dye. (B) Gel filtration chromatography reveals an Arp7–Arp9 heterodimer. Arp7–Arp9 complex eluted from Ni-NTA (lane 2) was combined with the size standards bovine serum albumin (BSA) and aldolase (lane 3) and separated on a Superdex-200 column. Samples were separated in a 7.5% acrylamide–SDS gel and visualized with Coomassie Blue.
Fig. 4. Arp7 and Arp9 are obligate partners in vivo. (A) Arp7 and Arp9 codependence for assembly into RSC or SWI/SNF complexes. Extracts were prepared from WT (YBC405), mra1 (YBC430), _mra1 arp9_Δ (YBC426) and _mra1 arp7_Δ (YBC427) strains (left panel). Immune complexes were formed with anti-Arp7 (middle panel) or anti-Arp9 (right panel), washed, eluted, immunoblotted and probed with anti-Sth1 or anti-Swi3 antiserum. Assembly defects of Arp9 in _arp7_Δ mutants in (B) RSC or (C) SWI/SNF. Whole-cell extracts from WT (YBC405), mra1 (YBC430) and _mra1 arp7_Δ (YBC427) were prepared (Loads). Immune complexes were formed with (B) anti-Sth1 or (C) anti-Swi3, washed, eluted, immunoblotted and probed with anti-Arp9 antiserum or antiserum specific for subunits of RSC (Rsc6) or SWI/SNF complex (Swp73) as controls. *Anti-Rsc6 and anti-Swp73 antibodies each cross react with a protein of slightly higher molecular weight. However, only the faster migrating species (bona fide Rsc6 or Swp73) is coprecipitated, providing an internal control for specificity.
Fig. 5. Composition and activity of RSC lacking Arp7 and Arp9. (A) Purification of RSCΔ7/9, a stable RSC complex lacking Arp7 or Arp9. RSC and RSCΔ7/9 were purified (see Materials and methods), and 600 ng of each was separated on a 7.5% acrylamide–SDS gel, followed by staining with silver. *A low-level degradation product of Sth1. (B) Immunoblot analysis of RSCΔ7/9. Purified RSC (125 ng) or RSCΔ7/9 (125 ng) was separated in a 7.5% acrylamide–SDS gel and immunoblotted with antiserum raised against Sth1, Rsc3, Rsc30, Rsc6, Arp7 or Arp9. (C) RSCΔ7/9 displaces a triple helix. A triple-helix substrate consisting of a 40-base triple-helical region was centered on a 190 bp duplex DNA, treated as indicated for 60 min at 30°C or heated briefly at 90°C (Heat) and separated in a 15% polyacrylamide gel. Displacement of the 32P-labeled third strand as a percentage of total (%Disp.) was quantified with Image Quant. (D) RSCΔ7/9 remodels mononucleosomes efficiently. Recombinant yeast octamers were assembled into mononucleosomes (172 bp 5S DNA, 32P-end labeled) containing a single _Dra_I site near the dyad and purified. Reaction components: RSC or RSCΔ7/9 (as indicated); nucleosomes (12 nM); ATP (1 mM); _Dra_I (20 units). Conditions: 1 h at 30°C, followed by DNA extraction, separation and autoradiography. (E) Reconstitution of the yeast Arp7/Arp9 heterodimer with RSCΔ7/9 and the requirement for the C-termini of Arp7 and Arp9 for assembly into RSC. Arp7/9 heterodimer derivatives were expressed and purified, normalized for Arp9 content, bound to anti-Flag M2 agarose, washed (200 mM NaCl), incubated with RSCΔ7/9 (4:1 molar ratio of Arp7/9 to RSC), washed (500 mM NaCl) and eluted with Flag peptide. Immunoblot analysis of RSC reconstitutions. L: load, 10% of total. E: eluate, 40%. (F) A recombinant (E.coli) Arp7/9 dimer reconstitutes into RSC. Purified Arp7/9 dimer from E.coli was bound to anti-Flag M2 agarose and treated as in (E). (G) Recombinant Arp7ΔC and Arp9ΔC copurify. Histidine-tagged Arp7ΔC (1–435) and Flag-tagged Arp9ΔC (1–437) were coexpressed in E.coli using a bi-cistronic vector and purified as described in Figure 3 and Materials and methods. Arp7ΔC and Arp9ΔC migrate at nearly the identical position.
Fig. 6. Multicopy NHP6A suppresses arp alleles. (A) Suppression of arp7 Ts– alleles by multicopy NHP6A. Plasmid YEplac195 NHP6A or empty vector were transformed into WT or the arp7 Ts– strains and grown at 28°C or 35°C. Strains: YBC726 transformants harboring WT pNCT.ARP7 or plasmids bearing arp7 Ts– alleles: pNCT.ARP7 (YBC1533), pNCT arp7E411K (YBC776) or pNCT.arp7S33F (YBC788). (B) Suppression of arp9 Ts– alleles by multicopy NHP6A. YEplac195 NHP6A plasmid or empty vector were transformed into WT or arp9 Ts– strains and compared for growth at 28°C or 37°C. Strains: YBC86 transformants harboring pNCT.ARP9 (YBC1535), pNCT.arp9-1(YBC1536) or pNCT. arp9_Δ_C (YBC775).
Fig. 7. RSC interacts with Nhp6a and facilitates its binding to nucleosomes. (A) Nhp6a co-immunoprecipitates Sth1. Immune complexes were formed with anti-Sth1 antibodies and whole-cell extract (from YBC1330, which expresses a chromosomal HA-tagged version of Nhp6a), treated with DNase I (30 units) (lanes 4 and 5; mock, lanes 2 and 3), washed, immunoblotted and probed with anti-HA antibody. Load, 2% of total; pellet, 100%. (B) RSC interacts with Nhp6a in the absence of Arp7 and Arp9. Whole-cell extract (1 mg) from an _arp7_Δ _arp9_Δ mra1 strain (YBC395) was incubated with conjugated anti-Nhp6 resin, treated with DNase I (30 U) (lanes 4 and 5; mock, lanes 2 and 3), washed (250 mM NaCl), immunoblotted, separated by SDS–PAGE and probed with anti-Sth1, anti-Rsc6 or anti-Nhp6. Load, 2% of total; pellet, 100%. (C) RSC loads Nhp6a onto nucleosomes. RSC (10 nM) was incubated with nucleosomes (1 nM) and Nhp6a (as indicated) for 20 min and resolved as described in Materials and methods. *The first species formed with Nhp6a only in the absence of ATP, which may represent binding to the exposed linker. (D) Nhp6a inhibits access to the _Dra_I site on nucleosomes. Nhp6a was titrated into remodeling reactions (see Figure 5D; Materials and methods).
Similar articles
- Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF.
Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Cairns BR, et al. Mol Cell. 1998 Nov;2(5):639-51. doi: 10.1016/s1097-2765(00)80162-8. Mol Cell. 1998. PMID: 9844636 - Structure of the full-length yeast Arp7-Arp9 heterodimer.
Lobsiger J, Hunziker Y, Richmond TJ. Lobsiger J, et al. Acta Crystallogr D Biol Crystallogr. 2014 Feb;70(Pt 2):310-6. doi: 10.1107/S1399004713027417. Epub 2014 Jan 29. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24531465 - Structural insights into assembly and function of the RSC chromatin remodeling complex.
Baker RW, Reimer JM, Carman PJ, Turegun B, Arakawa T, Dominguez R, Leschziner AE. Baker RW, et al. Nat Struct Mol Biol. 2021 Jan;28(1):71-80. doi: 10.1038/s41594-020-00528-8. Epub 2020 Dec 7. Nat Struct Mol Biol. 2021. PMID: 33288924 Free PMC article. - Assays for chromatin remodeling during nucleotide excision repair in Saccharomyces cerevisiae.
Zhang L, Jones K, Smerdon MJ, Gong F. Zhang L, et al. Methods. 2009 May;48(1):19-22. doi: 10.1016/j.ymeth.2009.03.017. Epub 2009 Mar 29. Methods. 2009. PMID: 19336254 Free PMC article. Review. - Electron microscopic analysis of the RSC chromatin remodeling complex.
Asturias FJ, Ezeokonkwo C, Kornberg RD, Lorch Y. Asturias FJ, et al. Methods Enzymol. 2004;376:48-62. doi: 10.1016/S0076-6879(03)76004-2. Methods Enzymol. 2004. PMID: 14975298 Review. No abstract available.
Cited by
- Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler.
Schubert HL, Wittmeyer J, Kasten MM, Hinata K, Rawling DC, Héroux A, Cairns BR, Hill CP. Schubert HL, et al. Proc Natl Acad Sci U S A. 2013 Feb 26;110(9):3345-50. doi: 10.1073/pnas.1215379110. Epub 2013 Feb 11. Proc Natl Acad Sci U S A. 2013. PMID: 23401505 Free PMC article. - The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer.
Hodges C, Kirkland JG, Crabtree GR. Hodges C, et al. Cold Spring Harb Perspect Med. 2016 Aug 1;6(8):a026930. doi: 10.1101/cshperspect.a026930. Cold Spring Harb Perspect Med. 2016. PMID: 27413115 Free PMC article. Review. - Sequence and comparative genomic analysis of actin-related proteins.
Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B. Muller J, et al. Mol Biol Cell. 2005 Dec;16(12):5736-48. doi: 10.1091/mbc.e05-06-0508. Epub 2005 Sep 29. Mol Biol Cell. 2005. PMID: 16195354 Free PMC article. - Actin complexes in the cell nucleus: new stones in an old field.
Castano E, Philimonenko VV, Kahle M, Fukalová J, Kalendová A, Yildirim S, Dzijak R, Dingová-Krásna H, Hozák P. Castano E, et al. Histochem Cell Biol. 2010 Jun;133(6):607-26. doi: 10.1007/s00418-010-0701-2. Epub 2010 May 5. Histochem Cell Biol. 2010. PMID: 20443021 Review. - Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast.
Monahan BJ, Villén J, Marguerat S, Bähler J, Gygi SP, Winston F. Monahan BJ, et al. Nat Struct Mol Biol. 2008 Aug;15(8):873-80. doi: 10.1038/nsmb.1452. Epub 2008 Jul 11. Nat Struct Mol Biol. 2008. PMID: 18622392 Free PMC article.
References
- Angus-Hill M.L., Schlichter,A., Roberts,D., Erdjument-Bromage,H., Tempst,P. and Cairns,B.R. (2001) A Rsc3–Rsc30 zinc cluster heterodimer reveals new roles for the chromatin remodeling complex RSC in gene expression and cell cycle control. Mol. Cell, 7, 741–751. - PubMed
- Boyer L.A. and Peterson,C.L. (2000) Actin-related proteins (Arps): conformational switches for chromatin- remodeling machines? BioEssays, 22, 666–672. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous