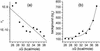Modeling of DNA microarray data by using physical properties of hybridization - PubMed (original) (raw)
Modeling of DNA microarray data by using physical properties of hybridization
G A Held et al. Proc Natl Acad Sci U S A. 2003.
Abstract
A method of analyzing DNA microarray data based on the physical modeling of hybridization is presented. We demonstrate, in experimental data, a correlation between observed hybridization intensity and calculated free energy of hybridization. Then, combining hybridization rate equations, calculated free energies of hybridization, and microarray data for known target concentrations, we construct an algorithm to compute transcript concentration levels from microarray data. We also develop a method for eliminating outlying data points identified by our algorithm. We test the efficacy of these methods by comparing our results with an existing statistical algorithm, as well as by performing a cross-validation test on our model.
Figures
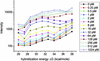
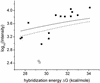
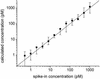
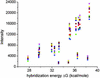
Fig. 1.
Observed hybridization intensity as a function of calculated hybridization free energy, –Δ_G_, for spike-in target concentrations between 0 and 1,024 pM. Each data point shows the average measured intensity for all probes with a calculated free energy that lies within a given energy bin at a particular target concentration. The centers of the 10 energy bins range between 26 and 38 kcal/mol, and their widths are all 1.33 kcal/mol. The average intensities are plotted at the values of the bin centers. An increase in observed hybridization intensity with increasing Δ_G_ is clearly observable. The lines simply connect the data points.
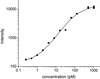
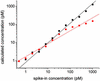
Fig. 2.
Observed hybridization intensity as a function of spike-in target concentration of PM probe one of gene 37777_at. Replicate measurements are shown as distinct data points. The solid line is a best fit of the data to Eq. 2, with _n_I, n e, and bg e adjustable parameters.
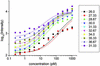
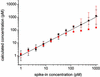
Fig. 3.
Observed hybridization intensity as a function of spike-in target concentration for the energy-binned data shown in Fig. 1. Units of energy are kcal/mol. Solid lines show the best fits of all the plotted data fit simultaneously to Eq. 2 (see Data Analysis).
Fig. 4.
Best-fit values of n e (a) and bg e (b) obtained from the fits shown in Fig. 3 vs. calculated Δ_G_. Solid lines are best fits to the plotted results (see Data Analysis).
Fig. 5.
Observed hybridization intensity as a function of calculated Δ_G_ for the probe set of gene 36311_at. The data were collected on a single GeneChip with a spike-in target concentration of 256 pM. The dotted line is a best fitof all the data to Eq. 2, with concentration c the only adjustable parameter; the best fit c is 92 pM. The solid line is a best fit to only the filled data points, the hollow ones having been identified as statistical outliers (see Data Analysis); the best fit c in this case is 151 pM, closer to the known spike-in value of 256 pM.
Fig. 6.
Median value of predicted concentrations of all transcript measurements taken at a given spike-in target concentration vs. that concentration. Results for the analysis discussed in the text and for Affymetrix (
microarray suite
- are plotted as black and red squares, respectively. The solid lines are best fits of the plotted results to power laws (see Analysis of the Accuracy of the Algorithm).
Fig. 7.
Range of predicted concentrations vs. spike-in target concentration. The error bars at each spike-in concentration show the range of predicted concentration values that must be encompassed to include half of the predicted values (see Analysis of the Accuracy of the Algorithm). The black and red error bars show ranges obtained by using nalysis described in the text and with Affymetrix (
microarray suite
5), respectively.
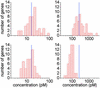
Fig. 8.
Histograms of concentration values predicted by our algorithm (Upper) and with Affymetrix (
microarray suite
5)(Lower) for all data at spike-in concentrations of 16 pM (Left) and 256 pM (Right).
Fig. 9.
Range of predicted concentrations obtained through the cross-validation procedure (see Analysis of the Accuracy of the Algorithm) vs. spike-in target concentration. The error bars are defined by the same criteria as in Fig. 7.
Fig. 10.
Observed hybridization intensity as a function of probe hybridization free energy for gene 1091_at at spike-in target concentration of 256 pM. Data points from different replicate measurements are shown in different colors.
Similar articles
- Physico-chemical foundations underpinning microarray and next-generation sequencing experiments.
Harrison A, Binder H, Buhot A, Burden CJ, Carlon E, Gibas C, Gamble LJ, Halperin A, Hooyberghs J, Kreil DP, Levicky R, Noble PA, Ott A, Pettitt BM, Tautz D, Pozhitkov AE. Harrison A, et al. Nucleic Acids Res. 2013 Mar 1;41(5):2779-96. doi: 10.1093/nar/gks1358. Epub 2013 Jan 9. Nucleic Acids Res. 2013. PMID: 23307556 Free PMC article. Review. - Secondary structure in the target as a confounding factor in synthetic oligomer microarray design.
Ratushna VG, Weller JW, Gibas CJ. Ratushna VG, et al. BMC Genomics. 2005 Mar 8;6:31. doi: 10.1186/1471-2164-6-31. BMC Genomics. 2005. PMID: 15755320 Free PMC article. - Modeling background intensity in DNA microarrays.
Kroll KM, Barkema GT, Carlon E. Kroll KM, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2008 Jun;77(6 Pt 1):061915. doi: 10.1103/PhysRevE.77.061915. Epub 2008 Jun 18. Phys Rev E Stat Nonlin Soft Matter Phys. 2008. PMID: 18643308 - Inverse Langmuir method for oligonucleotide microarray analysis.
Mulders GC, Barkema GT, Carlon E. Mulders GC, et al. BMC Bioinformatics. 2009 Feb 20;10:64. doi: 10.1186/1471-2105-10-64. BMC Bioinformatics. 2009. PMID: 19232092 Free PMC article. - [Properties of hybridization isotherms upon binding of ligands on microchips].
Golovkin MV, Matveeva OV, Nechipurenko IuD. Golovkin MV, et al. Biofizika. 2009 Sep-Oct;54(5):820-3. Biofizika. 2009. PMID: 19894619 Russian.
Cited by
- 16S rRNA gene-based oligonucleotide microarray for environmental monitoring of the betaproteobacterial order "Rhodocyclales".
Loy A, Schulz C, Lücker S, Schöpfer-Wendels A, Stoecker K, Baranyi C, Lehner A, Wagner M. Loy A, et al. Appl Environ Microbiol. 2005 Mar;71(3):1373-86. doi: 10.1128/AEM.71.3.1373-1386.2005. Appl Environ Microbiol. 2005. PMID: 15746340 Free PMC article. - Novel multi-nucleotide polymorphisms in the human genome characterized by whole genome and exome sequencing.
Rosenfeld JA, Malhotra AK, Lencz T. Rosenfeld JA, et al. Nucleic Acids Res. 2010 Oct;38(18):6102-11. doi: 10.1093/nar/gkq408. Epub 2010 May 20. Nucleic Acids Res. 2010. PMID: 20488869 Free PMC article. - Beyond Affymetrix arrays: expanding the set of known hybridization isotherms and observing pre-wash signal intensities.
Pozhitkov AE, Boube I, Brouwer MH, Noble PA. Pozhitkov AE, et al. Nucleic Acids Res. 2010 Mar;38(5):e28. doi: 10.1093/nar/gkp1122. Epub 2009 Dec 6. Nucleic Acids Res. 2010. PMID: 19969547 Free PMC article. - In situ-synthesized virulence and marker gene biochip for detection of bacterial pathogens in water.
Miller SM, Tourlousse DM, Stedtfeld RD, Baushke SW, Herzog AB, Wick LM, Rouillard JM, Gulari E, Tiedje JM, Hashsham SA. Miller SM, et al. Appl Environ Microbiol. 2008 Apr;74(7):2200-9. doi: 10.1128/AEM.01962-07. Epub 2008 Feb 1. Appl Environ Microbiol. 2008. PMID: 18245235 Free PMC article. - Physico-chemical foundations underpinning microarray and next-generation sequencing experiments.
Harrison A, Binder H, Buhot A, Burden CJ, Carlon E, Gibas C, Gamble LJ, Halperin A, Hooyberghs J, Kreil DP, Levicky R, Noble PA, Ott A, Pettitt BM, Tautz D, Pozhitkov AE. Harrison A, et al. Nucleic Acids Res. 2013 Mar 1;41(5):2779-96. doi: 10.1093/nar/gks1358. Epub 2013 Jan 9. Nucleic Acids Res. 2013. PMID: 23307556 Free PMC article. Review.
References
- Brown, P. O. & Botstein, D. (1999) Nat. Genet. 21, 33–37. - PubMed
- Lipshutz, R. J., Fodor, S. P. A., Gingeras, T. R. & Lockhart, D. J. (1999) Nat. Genet. 21, 20–24. - PubMed
- Lockhart, D. J., Dong, H. L., Byrne, M. C., Follettie, M. T., Gallo, M. V., Chee, M. S., Mittmann, M., Wang, C. W., Kobayashi, M., Horton, H. & Brown, E. L. (1996) Nat. Biotechnol. 14, 1675–1680. - PubMed
- Affymetrix (2001) Statistical Algorithms Reference Guide, Affymetrix Technical Note (Affymetrix, Santa Clara, CA).
- Mount, D. W. (2001) Bioinformatics: Sequence and Genome Analysis (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 519–523.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical